Bioinformatics explained: Protein statistics
Every protein holds specific and individual features which are unique to that particular protein. Features such as isoelectric point or amino acid composition can reveal important information of a novel protein. Many of the features described below are calculated in a simple way.
- Molecular weight
The molecular weight is the mass of a protein or molecule. The molecular weight is
simply calculated as the sum of the atomic mass of all the atoms in
the molecule.
The weight of a protein is usually represented in Daltons (Da).
A calculation of the molecular weight of a protein does not usually include additional post-translational modifications. For native and unknown proteins it tends to be difficult to assess whether posttranslational modifications such as glycosylations are present on the protein, making a calculation based solely on the amino acid sequence inaccurate. The molecular weight can be determined very accurately by mass-spectrometry in a laboratory.
- Isoelectric point
The isoelectric point (pI) of a protein is the pH where the proteins
has no net charge. The pI is calculated from the pKa values for
20 different amino acids. At a pH below the pI, the protein carries
a positive charge, whereas if the pH is above pI the proteins carry
a negative charge. In other words, pI is high for basic proteins and
low for acidic proteins. This information can be used in the
laboratory when running electrophoretic gels. Here the proteins can
be separated, based on their isoelectric point.
- Aliphatic index
The aliphatic index of a protein is a
measure of the relative volume occupied by aliphatic side chain of
the following amino acids: alanine, valine, leucine and isoleucine.
An increase in the aliphatic index increases the thermostability of
globular proteins. The index is calculated by the following formula.
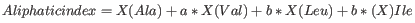
X(Ala), X(Val), X(Ile) and X(Leu) are the amino acid compositional fractions. The constants a and b are the relative volume of valine (a=2.9) and leucine/isoleucine (b=3.9) side chains compared to the side chain of alanine [Ikai, 1980].
- Estimated half-life
The half life of a protein is the time it takes
for the protein pool of that particular protein to be reduced to the
half. The half life of proteins is highly dependent on the presence
of the N-terminal amino acid, thus overall protein stability
[Bachmair et al., 1986,Gonda et al., 1989,Tobias et al., 1991]. The
importance of the N-terminal residues is generally known as the
'N-end rule'. The N-end rule and consequently the N-terminal amino
acid, simply determines the half-life of proteins. The estimated
half-life of proteins have been investigated in mammals, yeast and
E. coli (see the table
below). If leucine is found N-terminally in mammalian proteins the
estimated half-life is 5.5 hours.
Amino acid Mammalian Yeast E. coli Ala (A) 4.4 hour >20 hours >10 hours Cys (C) 1.2 hours >20 hours >10 hours Asp (D) 1.1 hours 3 min >10 hours Glu (E) 1 hour 30 min >10 hours Phe (F) 1.1 hours 3 min 2 min Gly (G) 30 hours >20 hours >10 hours His (H) 3.5 hours 10 min >10 hours Ile (I) 20 hours 30 min >10 hours Lys (K) 1.3 hours 3 min 2 min Leu (L) 5.5 hours 3 min 2 min Met (M) 30 hours >20 hours >10 hours Asn (N) 1.4 hours 3 min >10 hours Pro (P) >20 hours >20 hours ? Gln (Q) 0.8 hour 10 min >10 hours Arg (R) 1 hour 2 min 2 min Ser (S) 1.9 hours >20 hours >10 hours Thr (T) 7.2 hours >20 hours >10 hours Val (V) 100 hours >20 hours >10 hours Trp (W) 2.8 hours 3 min 2 min Tyr (Y) 2.8 hours 10 min 2 min - Extinction coefficient
This measure indicates how much light
is absorbed by a protein at a particular wavelength. The extinction
coefficient is measured by UV spectrophotometry, but can also be
calculated. The amino acid composition is important when calculating
the extinction coefficient. The extinction coefficient is calculated
from the absorbance of cysteine, tyrosine and tryptophan.
Two values are reported. The first value, "Non-reduced cysteines", is computed assuming that all cysteine residues appear as half cystines, meaning they form di-sulfide bridges to other cysteines:
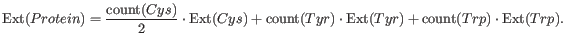
The second value, "Reduced cysteines", assumes that no di-sulfide bonds are formed:
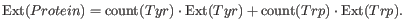
The extinction coefficient values of the three important amino acids at different wavelengths are found in [Gill and von Hippel, 1989] or in [Pace et al., 1995]. At 280nm the extinction coefficients are
- [Gill and von Hippel, 1989]: Ext(Cystine) = 120, Ext(Tyr) = 1280 and Ext(Trp) = 5690
- [Pace et al., 1995]: Ext(Cystine) = 125, Ext(Tyr) = 1490 and Ext(Trp) = 5500
This equation is only valid under the following conditions:
- pH 6.5
- 6.0 M guanidium hydrochloride
- 0.02 M phosphate buffer
Knowing the extinction coefficient, the absorbance (optical density) can be calculated using the following formula:
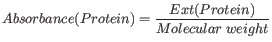
- Atomic composition Amino acids are indeed very simple compounds. All 20 amino acids consist of combinations of only five different atoms. The atoms which can be found in these simple structures are: Carbon, Nitrogen, Hydrogen, Sulfur, Oxygen. The atomic composition of a protein can for example be used to calculate the precise molecular weight of the entire protein.
- Total number of negatively charged residues (Asp + Glu) At neutral pH, the fraction of negatively charged residues provides information about the location of the protein. Intracellular proteins tend to have a higher fraction of negatively charged residues than extracellular proteins.
- Total number
of positively charged residues (Arg + Lys) At neutral pH, nuclear
proteins have a high relative percentage of positively charged amino
acids. Nuclear proteins often bind to the negatively charged DNA,
which may regulate gene expression or help to fold the DNA. Nuclear
proteins often have a low percentage of aromatic residues
[Andrade et al., 1998].
- Amino acid distribution
Amino acids are the basic components
of proteins. The amino acid distribution in a protein is simply the
percentage of the different amino acids represented in a particular
protein of interest. Amino acid composition is generally conserved
through family-classes in different organisms which can be useful
when studying a particular protein or enzymes across species
borders. Another interesting observation is that amino acid
composition variate slightly between proteins from different
subcellular localizations. This fact has been used in several
computational methods, used for prediction of subcellular
localization.
- Annotation table
This table provides an overview of all the
different annotations associated with the sequence and their
incidence.
- Dipeptide distribution This measure is simply a count, or frequency, of all the observed adjacent pairs of amino acids (dipeptides) found in the protein. It is only possible to report neighboring amino acids. Knowledge on dipeptide composition have previously been used for prediction of subcellular localization.
