TaqMan
CLC Genomics Workbench allows the user to design primers and probes for TaqMan PCR applications.
TaqMan probes are oligonucleotides that contain a fluorescent reporter dye at the 5' end and a quenching dye at the 3' end. Fluorescent molecules become excited when they are irradiated and usually emit light. However, in a TaqMan probe the energy from the fluorescent dye is transferred to the quencher dye by fluorescence resonance energy transfer as long as the quencher and the dye are located in close proximity i.e. when the probe is intact. TaqMan probes are designed to anneal within a PCR product amplified by a standard PCR primer pair. If a TaqMan probe is bound to a product template, the replication of this will cause the Taq polymerase to encounter the probe. Upon doing so, the 5'exonuclease activity of the polymerase will cleave the probe. This cleavage separates the quencher and the dye, and as a result the reporter dye starts to emit fluorescence.
The TaqMan technology is used in Real-Time quantitative PCR. Since the accumulation of fluorescence mirrors the accumulation of PCR products it can can be monitored in real-time and used to quantify the amount of template initially present in the buffer.
The technology is also used to detect genetic variation such as SNP's. By designing a TaqMan probe which will specifically bind to one of two or more genetic variants it is possible to detect genetic variants by the presence or absence of fluorescence in the reaction.
A specific requirement of TaqMan probes is that a G nucleotide can not be present at the 5' end since this will quench the fluorescence of the reporter dye. It is recommended that the melting temperature of the TaqMan probe is about 10 degrees celsius higher than that of the primer pair.
Primer design for TaqMan technology involves designing a primer pair and a TaqMan probe.
In TaqMan the user must thus define three regions: a Forward primer region, a Reverse primer region, and a TaqMan probe region. The easiest way to do this is to designate a TaqMan primer/probe region spanning the sequence region where TaqMan amplification is desired. This will automatically add all three regions to the sequence. If more control is desired about the placing of primers and probes the Forward primer region, Reverse primer region and TaqMan probe region can all be defined manually. If areas are known where primers or probes must not bind (e.g. repeat rich areas), one or more No primers here regions can be defined. The regions are defined by making a selection on the sequence and right-clicking the selection.
It is required that at least a part of the Forward primer region is located upstream of the TaqMan Probe region, and that the TaqMan Probe region, is located upstream of a part of the Reverse primer region.
In TaqMan mode the Inner melting temperature menu in the primer parameters panel is activated allowing the user to set a separate melting temperature interval for the TaqMan probe.
After exploring the available primers (see Graphical display of primer information) and setting the desired parameter values in the Primer Parameters preference group, the Calculate button will activate the primer design algorithm.
After pressing the Calculate button a dialog will appear (see figure 19.10) which is similar to the Nested PCR dialog described above (see Nested PCR dialog).
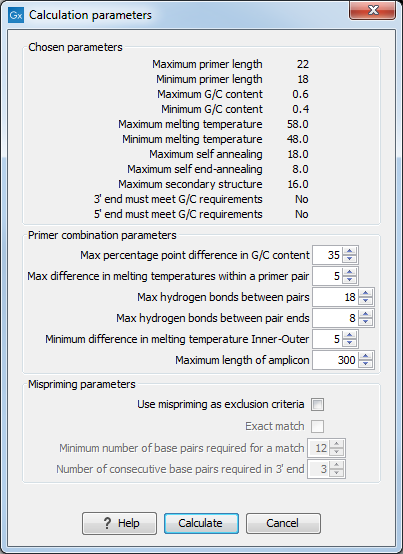
Figure 19.10: Calculation dialog for taqman primers.
In this dialog the options to set a minimum and a desired melting temperature difference between outer and inner refers to primer pair and probe respectively.
Furthermore, the central part of the dialog contains an additional parameter
- Maximum length of amplicon - determines the maximum length of the PCR fragment generated in the TaqMan analysis.
TaqMan output table In TaqMan mode there are two primers and a probe in a given solution, forward primer (F), reverse primer (R) and a TaqMan probe (TP).
The output table can show primer/probe-pair combination parameters for all three combinations of primers and single primer parameters for both primers and the TaqMan probe (see section on Standard PCR for an explanation of the available primer-pair and single primer information).
The fragment length in this mode refers to the length of the PCR fragment generated by the primer pair, and this is also the PCR fragment which can be exported.
