PacBio
Choosing the PacBio importer will open the dialog shown in figure 7.13. This data type can also be imported using the on-the-fly import functionality described in Launching workflows individually and in batches.
Note: PacBio HiFi reads are not fully supported. Of particular note: Tools that carry out de novo assembly do not support this data type, and quality scores are capped at 64, affecting tools that make use of such scores, e.g. variant detection tools.
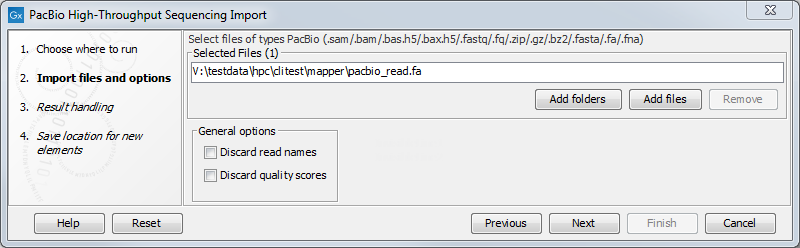
Figure 7.13: Importing data from PacBio.
We support import of the following file formats containing PacBio reads:
- H5 files (.bas.h5/.bax.h5) which contain one of two things. .bas.h5 files produced by instruments prior to PacBio RS II contain sequencing data such as reads and quality scores. .bas.h5 files from more recent PacBio instruments contain a list of .bax.h5 files where the actual sequencing data is stored. When importing H5 files, the user needs to select both the .bas.h5 file and all the accompanying .bax.h5 files belonging to a data set.
- Fastq files (.fastq) which contain sequence data and quality scores. Compressed Fastq (.fastq.gz) files are also supported.
- Fasta files (.fasta) which contain sequence data. Compressed Fasta (.fasta.gz) files are also supported.
- SAM or BAM files (.sam/.bam). The mapping information is discarded during import.
Under General options you have the following choices:
- Discard read names. For high-throughput sequencing data, the naming of the individual reads is often irrelevant given the huge amount of reads. This option allows you to discard read names to save disk space.
- Discard quality scores. Quality scores can be visualized in the mapping view and used for SNP detection. If this is not relevant for your work, you can choose to Discard quality scores. Discarding quality scores will reduce both disk space usage and memory consumption. As PacBio quality scores currently contain very little information, we recommend that you discard them. When importing Fasta files, this option is not available, since Fasta files do not contain quality scores.
Click Next and choose how the result of the import should be handled. We recommend choosing Save which will save the results directly to the disk.
When opening the "Element info" of sequence lists imported with the PacBio importer, the item "Platform" will display the mention PACBIO. For PacBio reads imported without the PacBio importer, it is possible to edit that field to "PACBIO" by clicking Edit next to the "Read Group" section in the Element Info view. Having the platform set to PacBio will ensure that the read mapper will perform better on PacBio reads.
