Explore Novel miRNAs
The Quantify miRNA tool only finds and quantifies the expression of known miRNAs. Here we will describe an approach to exploring novel miRNAs in the data.
First, take the "Unmapped reads" output from Quantify miRNA and use these reads as input to
Map Reads to Reference (![]() ).
To home in on sequences with a decent amount of expression and avoid sequences that map as a result of sequencing errors,
we can use the tools
Create Mapping Graph (
).
To home in on sequences with a decent amount of expression and avoid sequences that map as a result of sequencing errors,
we can use the tools
Create Mapping Graph (![]() )
and
Identify Graph Treshold Areas (
)
and
Identify Graph Treshold Areas (![]() )
to only consider sequences expressed above a user defined level.
)
to only consider sequences expressed above a user defined level.

Figure 31.64: Region in the human reference genome, with high coverage of reads that could not map to the miRNA reference database.
Next, to extract the sequence in the regions of high coverage use the tool
Extract Annotations (![]() ).
).
Finally, the tool
Predict Secondary Structure (![]() )
can be used to check if the structure matches the expected hairpin structure of miRNA precursors.
)
can be used to check if the structure matches the expected hairpin structure of miRNA precursors.
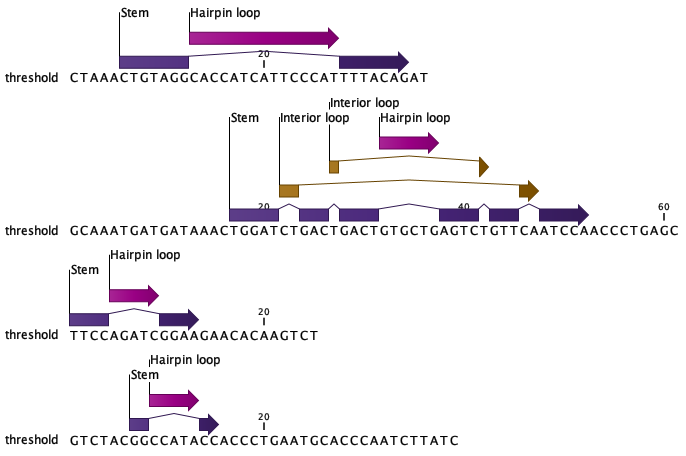
Figure 31.65: Sequences extracted from regions with high coverage and annotated with predicted secondary structure.
