Tool output and GAF file comparison
It can happen that you will find some discrepancy in the number of genes in your Gene Set test results and the original GAF file. This is also the case for results of the Hypergeometric Tests on Annotations, and Gene Set Enrichment Analysis (GSEA) tools.
In some cases, the result contains more genes than expected for a GO term. When testing for the significance of a particular GO term, we take into account that GO has a hierarchical structure. For example, when testing for the term "GO:0006259 DNA metabolic process", we include all genes that are annotated with more specific GO terms that are types of DNA metabolic process. As can be seen on figure 30.44, these include genes that are annotated with the more specific term "GO:0033151 V(D)J recombination". This is because "GO:0033151 V(D)J recombination" is a subtype of "GO:0002562 somatic diversification of immune receptors via germline recombination within a single locus", which in turn is a subtype of "GO:0016444 somatic diversification of immune receptors", which is a subtype of "GO:0006310 DNA recombination", which is a subtype of the original search term "GO:0006259 DNA metabolic process". Websites like geneontology.org ([Ashburner et al., 2000] and [The Gene Ontology Consortium, 2019]) provide an overview of the hierarchical structure of GO annotations.
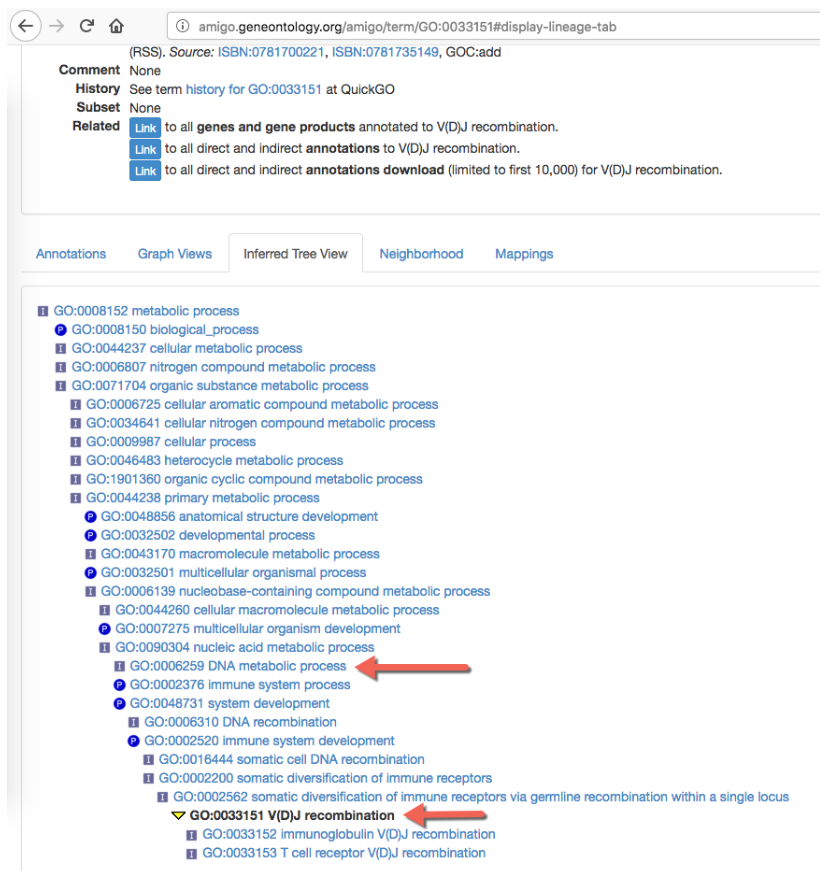
Figure 30.44: Hierarchical structure of GO annotations from geneontology.org.
In other cases, some annotations in the GAF file are missing from the Gene Set Test result. If the option "Exclude computationally inferred GO terms" is selected, then annotations in the GAF file that are computationally inferred (their description includes the [IEA] tag as in figure 30.45) will be excluded from the result. Thus, if the GAF file shows that almost all annotations are computationally inferred, it is recommended to run the tool without "Exclude computationally inferred GO terms".
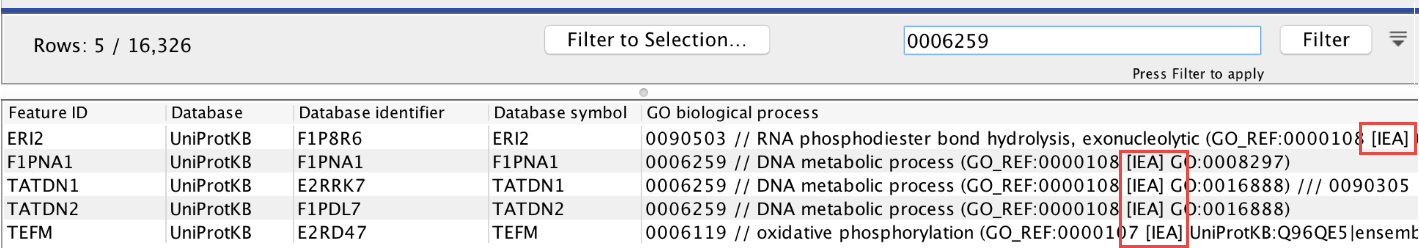
Figure 30.45: The [IEA] tag describes annotations that are computationally inferred.
