Import high-throughput sequencing data
CLC Genomics Workbench has dedicated tools for importing data from the following High-throughput sequencing systems:
- QIAGEN GeneReader
- Illumina Genome Analyzer, Nextseq, HiSeq and MiSeq
- PacBio
- Ion Torrent
Sequencing data from these systems, as well as Sanger and Fasta format files, can be imported using dedicated tools. Alternatively, this data can be imported using the on-the-fly functionality available in workflows, described in Launching workflows individually and in batches.
Importing other NGS related formats
- There are dedicated NGS importers for Sanger or Fasta format data.
- There is a dedicated import tool for read mappings in SAM/BAM format. Alignments of Complete Genomics data can be imported using this.
- An importer for Roche 454 sequencing data is available in the Legacy Tools folder.
- Complete Genomics master VAR files can be converted to VCF using tools provided by Complete Genomics, and imported into the CLC Genomics Workbench using the VCF track importer.
Once imported, data originating from any sequencing platform can be analyzed in the CLC Genomics Workbench.
Clicking on the Import (![]() ) button in the top toolbar will bring up a list of the supported data types as shown in figure 6.7. Select the appropriate format to launch the importer.
) button in the top toolbar will bring up a list of the supported data types as shown in figure 6.7. Select the appropriate format to launch the importer.
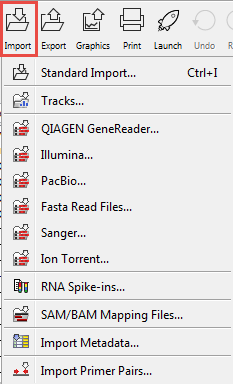
Figure 6.7: Choosing what kind of data you wish to import.
To specify the files to import, select either Add folders, in which case you then choose one or several folders from which all the files should be imported, or Add files, in which case you select individual files to import. Once files have been selected, configure the import options, which are described in the following sections.
Files can be removed from the list by selecting them and clicking on the Remove button.
If the wrong NGS importer was used to import your data, please check, and edit if necessary, the "Read Group" information in the "Element Info" view. To edit this information, choose from the drop-down menu the sequencing platform used to generate the data (figure 6.8) and click OK.
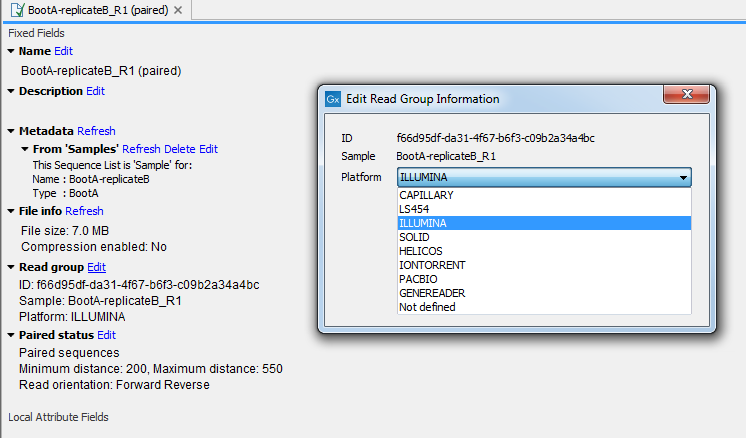
Figure 6.8: Editing the platform used to generate the data in the "Element Info" view.
Subsections
- QIAGEN GeneReader
- Illumina
- PacBio
- Fasta read files
- Sanger sequencing data
- Ion Torrent
- General notes on handling paired data
- SAM and BAM mapping files
