Differential Expression in Two Groups
The Differential Expression in Two Groups tool performs a statistical differential expression test for a set of Expression Tracks and a control. It uses multi-factorial statistics based on a negative binomial GLM as described in The statistical model. Differential Expression in Two Groups only handles one factor and two groups, as opposed to the Differential Expression for RNA-Seq tool that can handle multiple factors and multiple groups.
To run the Differential Expression in Two Groups analysis:
Toolbox | RNA-Seq and Small RNA Analysis (![]() )| Differential Expression in Two Groups (
)| Differential Expression in Two Groups (![]() )
)
In the first dialog (figure 30.22), select a number of Expression tracks (![]() ) (GE or TE) and click Next.
For Transcripts Expression Tracks (TE), the values used as input are "Total transcript reads".
For Gene Expression Tracks (GE), the values used depend on whether an eukaryotic or prokaryotic organism is analyzed, i.e., if the option "Genome annotated with Genes and transcripts" or "Genome annotated with Genes only" was used. For Eukaryotes the values are "Total Exon Reads", whereas for Prokaryotes the values are "Total Gene Reads".
) (GE or TE) and click Next.
For Transcripts Expression Tracks (TE), the values used as input are "Total transcript reads".
For Gene Expression Tracks (GE), the values used depend on whether an eukaryotic or prokaryotic organism is analyzed, i.e., if the option "Genome annotated with Genes and transcripts" or "Genome annotated with Genes only" was used. For Eukaryotes the values are "Total Exon Reads", whereas for Prokaryotes the values are "Total Gene Reads".
Note that the tool can be run in batch mode, albeit with the same control group expression for all selected batch units.
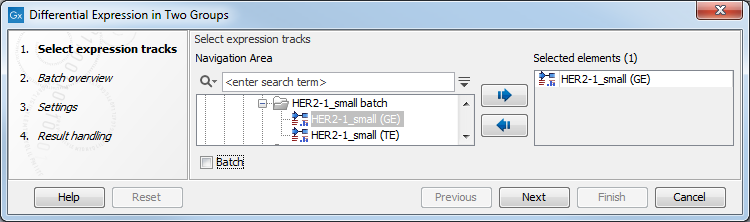
Figure 30.22: Select expression tracks for analysis.
In the Settings dialog, select a number of control Expression tracks (![]() ) (GE or TE). A warning message (as seen in figure 30.23) appears if only one track is selected for either the input or the control group: such a setting does not provide replicates, thus does not ensure sufficient statistical power to the analysis.
) (GE or TE). A warning message (as seen in figure 30.23) appears if only one track is selected for either the input or the control group: such a setting does not provide replicates, thus does not ensure sufficient statistical power to the analysis.
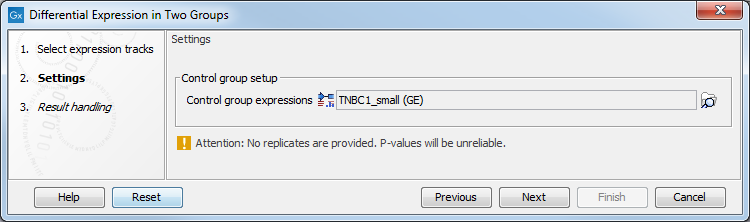
Figure 30.23: Select enough control expression tracks to ensure that replicates are provided.
The output of the tool is a comparison table study vs. control that can be visualized as a Statistical comparison track and a Volcano plot.
