Advanced options
The free energy minimization algorithm
includes a number of advanced options:
- Avoid isolated base pairs. The algorithm filters out isolated base pairs (i.e. stems of length 1).
- Apply different energy rules for Grossly Asymmetric Interior Loops (GAIL). Compute the minimum free energy applying different rules for Grossly Asymmetry Interior Loops (GAIL).
A Grossly Asymmetry Interior Loop (GAIL) is an interior loop that is
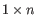 or
or 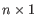 where
where 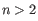 (see http://mfold.rna.albany.edu/doc/mfold-manual/node5.php).
(see http://mfold.rna.albany.edu/doc/mfold-manual/node5.php).
- Include coaxial stacking energy rules. Include free energy increments of coaxial stacking for adjacent helices [Mathews et al., 2004].
- Apply base pairing constraints. With base pairing constraints, you can easily add experimental constraints to your folding algorithm.
When you are computing suboptimal structures, it is not possible to
apply base pair constraints. The possible base pairing constraints
are:
- Force two equal length intervals to form a stem.
- Prohibit two equal length intervals to form a stem.
- Prohibit all nucleotides in a selected region to be a part of a base pair.
- Maximum distance between paired bases. Forces the algorithms to only consider RNA structures of a given upper length by setting a maximum distance between the base pair that opens a structure.
Specifying structure constraints
Structure constraints can serve two purposes in CLC Genomics Workbench: they can act as experimental constraints imposed on the MFE structure prediction algorithm or they can form a structure hypothesis to be evaluated using the partition function.
To force two regions to form a stem, open a normal sequence view and:
Select the two regions you want to force by pressing
Ctrl while selecting - (use ![]() on Mac) | right-click the
selection | Add Structure Prediction Constraints|
Force Stem Here
on Mac) | right-click the
selection | Add Structure Prediction Constraints|
Force Stem Here
This will add an annotation labeled "Forced Stem" to the sequence (see figure 23.5).
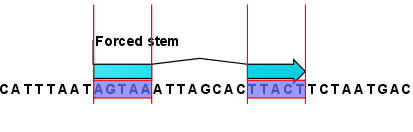
Figure 23.5: Force a stem of the selected bases.
Using this procedure to add base pairing constraints will force the algorithm to compute minimum free energy and structure with a stem in the selected region. The two regions must be of equal length.
To prohibit two regions to form a stem, open the sequence and:
Select the two regions you want to prohibit by
pressing Ctrl while selecting - (use ![]() on Mac) |
right-click the selection | Add Structure Prediction
Constraints | Prohibit Stem Here
on Mac) |
right-click the selection | Add Structure Prediction
Constraints | Prohibit Stem Here
This will add an annotation labeled "Prohibited Stem" to the sequence (see figure 23.6).
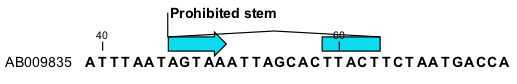
Figure 23.6: Prohibit the selected bases from forming a stem.
Using this procedure to add base pairing constraints will force the algorithm to compute minimum free energy and structure without a stem in the selected region. Again, the two selected regions must be of equal length.
To prohibit a region to be part of any base pair, open the sequence and:
Select the bases you don't want to base pair | right-click the selection | Add Structure Prediction Constraints | Prohibit From Forming Base Pairs
This will add an annotation labeled "No base pairs" to the sequence, see 23.7.
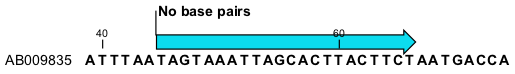
Figure 23.7: Prohibiting any of the selected base from pairing with other bases.
Using this procedure to add base pairing constraints will force the algorithm to compute minimum free energy and structure without a base pair containing any residues in the selected region.
When you click Predict secondary
structure (![]() ) and click Next, check
Apply base pairing constraints in order to force or
prohibit stem regions or prohibit regions from forming base pairs.
) and click Next, check
Apply base pairing constraints in order to force or
prohibit stem regions or prohibit regions from forming base pairs.
You can add multiple base pairing constraints, e.g. simultaneously adding forced stem regions and prohibited stem regions and prohibit regions from forming base pairs.
