Output from the Perform QIAseq RNA Fusion XP Analysis workflow
The Perform QIAseq RNA Fusion XP Analysis workflow's main outputs (figure 15.8) are two Genome Browser Views, a Gene Expression track, a Fusion Report and a Fusion track (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes_tool.html), and a Combined QC report that contains summary information from reports output by multiple tools in the workflow.
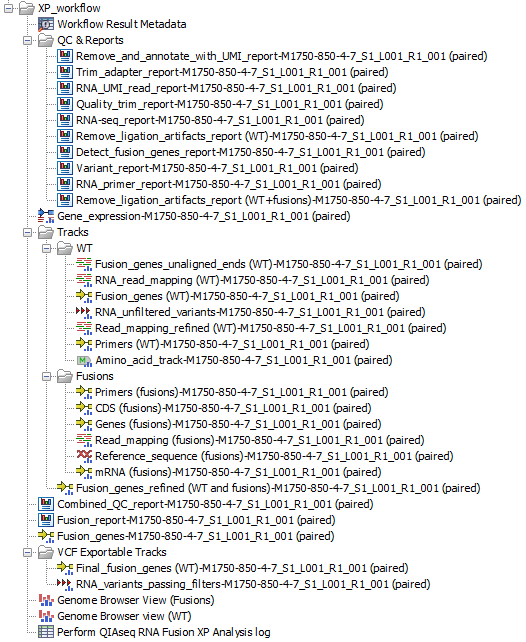
Figure 15.8: Outputs of the Perform QIAseq RNA Fusion XP Analysis workflow for an input called "M1750-850-4-7_S1_L001_R1_001 (paired)"
Genome Browser View (WT) displays the following tracks:
- Reference sequence tracks
- Reference gene tracks
- Reference mRNA tracks
- Reference CDS tracks
- The Fusion panel primers track
- An amino acid track that displays a graphical representation of the amino acid changes. The track is based on the CDS track and in addition to the amino acid sequence of the coding sequence, all amino acids that have been affected by variants are shown as individual amino acids below the amino acid track. Changes causing a frameshift are symbolized with two arrow heads, and variants causing premature stop are marked with an asterisk.
- A Variants Track of RNA variants passing filters
- The detected fusion genes track (based on the wild type genome sequence as reference)
- The mapping of the UMI reads
Genome Browser View (Fusions) displays the following tracks:
- Fusion sequence tracks (including an artificial intronic sequence of 50 N nucleotides).
- Fusion gene tracks
- Fusion mRNA tracks
- Fusion CDS tracks: each fusion CDS includes a "Frame aligns" annotation, which shows whether the CDS stays in the reading frame across the fusion breakpoint, allowing users to visualize fusions that are more likely to be translated to protein.
- The Fusion panel primers track
- The detected fusion genes track (based on a fusion genome sequence as reference)
- The mapping of the UMI reads
Many of the track outputs included in the Genome Browser Views are placed in two folders: "Tracks | WT" and "Tracks | Fusions".
Detailed reports that are summarized in the Combined QC report are placed in a folder called "QC & Reports". These include:
- A Remove and Annotate with UMI Report
- A Trim Adapters Report (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Trim_output.html)
- An RNA primer report described in The QIAseq RNAscan Panels Report.
- A Refined Fusion Gene report (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes_tool.html)
The VCF Exportable Tracks folder contains outputs that can be exported together as a single VCF file using the VCF exporter. This folder contains a Variant Track of RNA variants passing filters, and a track of fusions.
A second Variant Track of RNA unfiltered variants is found in the "Tracks | WT" folder. This is provided so that you can also review why a variant that was expected in the output has been filtered out of the RNA variants passing filters track.
The difference between the RNA unfiltered variant track and the RNA variants passing filters track depends on the following options available in the filtering steps:
- Filter based on quality criteria: Average Quality (quality of the sequenced bases that carry the variant), QUAL (significance of the variant), Read Position Test Probability (relative location of the variant in the reads that cover the variant position) and Read Direction Test Probability (relative presence of the variant in the reads from different directions that cover the variant position).
- Remove homopolymer error type variants, i.e., errors of the indel type that occur in homopolymer regions. These regions are known to be harder to sequence than non-homopolymeric regions.
- Remove false positive based on frequency The variant's frequency needs to be above that threshold for the variant to be output by the workflow in the filtered variant track. Note that the unfiltered variant track is generated by the Low Frequency Variant Detection tool run with a frequency cut-off value of 2.5. This value can be considered a pre-filter, which is initially applied to each site in the alignment and determines which sites the variant caller should consider potential variant sites when it starts the error rate and site type/frequencies parameter estimation. In the case of this option, a frequency cut-off is applied on the final candidate variant set (after variants that span across multiple alignment sites have been reconstructed). It is only meaningful to apply this post-filter at a value that is at least as high as the pre-filter value, and we actually recommend using a value that is as least twice as high (5.0). This allows for some wiggle-room when going from the single-site to the multiple site variant construction, in particular to avoid that long indels are fragmented due to coverage difference throughout the considered region.
For quality control of fusion calls, see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Interpretation_fusion_results.html. We particularly recommend carrying out manual quality control checks on results that include fusions with novel exon boundaries.
