Output from the Detect QIAseq RNAscan Fusions workflow
The Detect QIAseq RNAscan Fusions workflow's main outputs are two Genome Browser Views, a Gene Expression track, a Transcript Expression track, and a Combined QC report that contains summary information from reports output by multiple tools in the workflow.
Genome Browser View (WT) displays the following tracks:
- Reference mRNA tracks
- Reference gene tracks
- Reference sequence tracks
- The Fusion panel primers track
- The detected fusion genes track (based on the wild type genome sequence as reference)
- The mapping of the UMI reads
Genome Browser View (Fusions) displays the following tracks:
- The detected fusion genes track (based on a fusion genome sequence as reference)
- The mapping of the UMI reads
- Fusion sequence tracks (including an artificial intronic sequence of 50 N nucleotides).
- Fusion mRNA tracks
- Fusion gene tracks
- Fusion CDS tracks: each fusion CDS includes a "Frame aligns" annotation, which shows whether the CDS stays in the reading frame across the fusion breakpoint, allowing users to visualize fusions that are more likely to be translated to protein.
- The Fusion panel primers track
Many of the track outputs included in the Genome Browser Views are placed in two folders: "Tracks" and "Fusion References".
Detailed reports that are summarized in the Combined QC report are placed in a folder called "QC & Reports". These include:
- A QC for Sequencing Reads Graphical Report (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=QC_Sequencing_Reads.html)
- A Remove and Annotate with UMI Report
- A Trim Adapters Report (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Trim_output.html)
- A QC for RNAscan Panels Report described in The QIAseq RNAscan Panels Report.
- A Fusion Gene report (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes_tool.html)
The VCF Exportable Tracks folder contains a track of fusions that can be exported to VCF format using the VCF exporter.
Reviewing the results
The easiest way to review the results is to open the Genome Browser Views, as shown in figure 15.4.
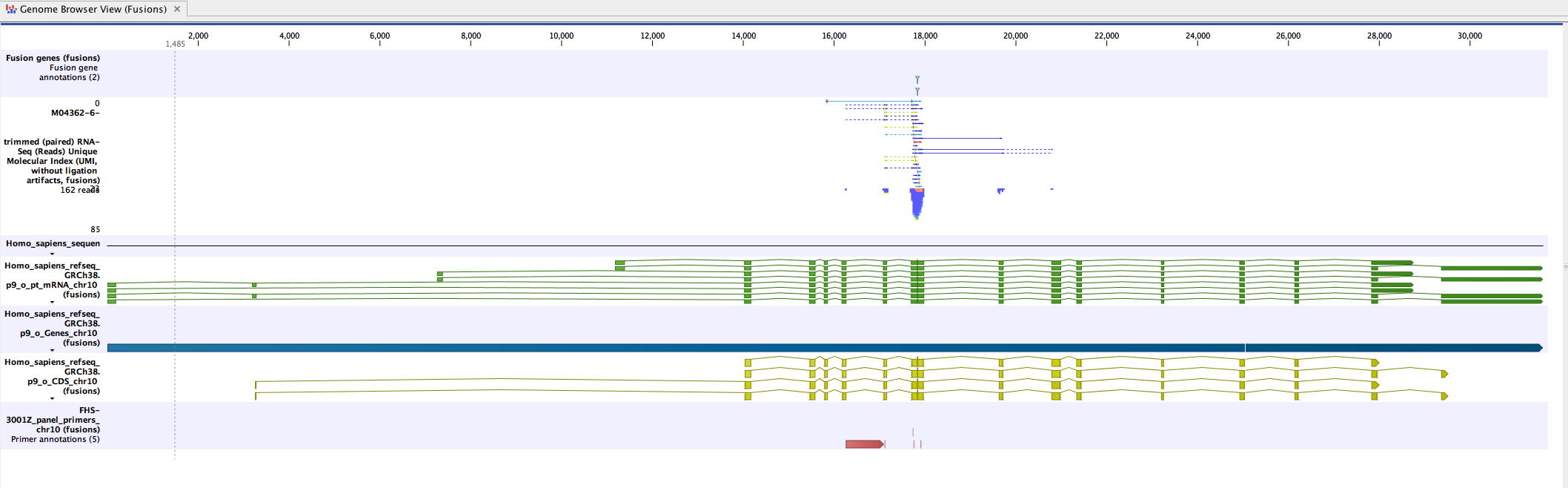
Figure 15.4: An example of a fusion in a Genome Browser View.
Double-click on the fusion track name (to the right of the Genome Browser View) while pressing the Ctrl button. The fusion track will open as a table in split view, below the Genome Browser View. Clicking on a fusion event in the table will zoom in to its location in the UMI read mapping, allowing you to review the UMI reads supporting the detected fusion.
Each line in the table corresponds to a fusion breakpoint, such that a fusion event is represented by two lines in the table. The two lines are linked by sharing the same "Fusion number", which identifies the fused genes, and the same "Fusion pair", which identifies the event for the gene. For more details on the table, see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes_tool.html.
For quality control of fusion calls, see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Interpretation_fusion_results.html. We particularly recommend carrying out manual quality control checks on results that include fusions with novel exon boundaries.
When running QIAseq Targeted RNAscan Panels, it is expected to detect HALC1-COLQ and TMP3-TMP4 (TMP4-TMP3) as false positive fusions; additional false positive fusions may be found when running custom made panels.
