Identify QIAseq xHYB Germline Variants including Mitochondrial
The Identify QIAseq xHYB Germline Variants including Mitochondrial template workflow calls germline variants in QIAseq xHYB panel target regions, as described in Identify QIAseq xHYB Germline Variants and in target regions associated with a mitochondrial spike-in.
It can be found at:
Template Workflows | Biomedical Workflows (![]() ) | QIAseq Sample Analysis (
) | QIAseq Sample Analysis (![]() ) | QIAseq DNA workflows (
) | QIAseq DNA workflows (![]() ) | Identify QIAseq xHYB Germline Variants including Mitochondrial (
) | Identify QIAseq xHYB Germline Variants including Mitochondrial (![]() )
)
This workflow can also be launched from the Analyze QIAseq Samples guide, which is described in The Analyze QIAseq Samples guide. It is available under the xHYB Human tab.
Mitochondrial variant calling is performed using the Low Frequency Variant Detection tool. The parameters for this can be set in the relevant wizard step when launching the workflow (figure 14.58). For a description of the different parameters that can be adjusted, see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Low_Frequency_Variant_Detection.html. If you click on "Locked Settings", you will be able to see all parameters used for variant detection in the template workflow. Resulting variants are filtered using a different filtering cascade compared to variants obtained from the Fixed Ploidy Variant Detection tool. The settings of the filtering cascade are locked by default.
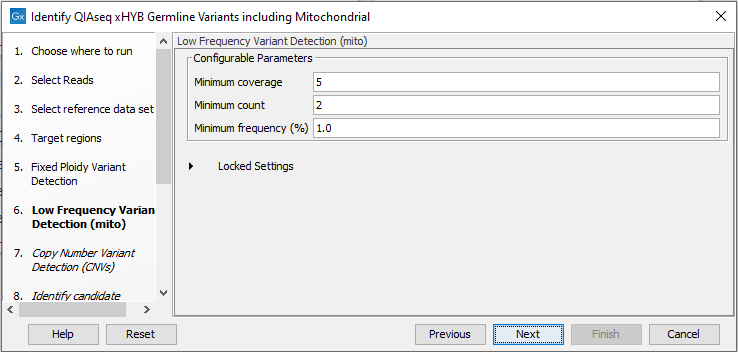
Figure 14.58: Specify the parameters for the Low Frequency Variant Detection tool.
Please note that copy number variation analysis is restricted to QIAseq xHYB panel target regions and will therefore not be performed for mitochondrial target regions.
The following output files are created separately for mitochondrial data:
- A Coverage report (
 ) and a Per region coverage track (
) and a Per region coverage track ( ) from the QC for Target Sequencing tool (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=QC_Targeted_Sequencing.html).
) from the QC for Target Sequencing tool (see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=QC_Targeted_Sequencing.html).
- Three variant tracks (
 ): Two from the Low Frequency Variant Detection tool: the Unfiltered Variants is output before the filtering steps, the Variants passing filters is the one used in the Genome Browser View (see
http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=_annotated_variant_table.html for a definition of the variant table content). The third is the Indels indirect evidence track produced by the Structural Variant Caller. This is also available in the Genome Browser View.
): Two from the Low Frequency Variant Detection tool: the Unfiltered Variants is output before the filtering steps, the Variants passing filters is the one used in the Genome Browser View (see
http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=_annotated_variant_table.html for a definition of the variant table content). The third is the Indels indirect evidence track produced by the Structural Variant Caller. This is also available in the Genome Browser View.
- An Amino acid track (
 ) generated using the mitochondrial genetic code.
) generated using the mitochondrial genetic code.
