WGS, WES, TAS and WTS template workflow descriptions
CLC Workbench contains several template workflows that support analysis of cancer data, but also analysis of hereditary diseases and other conditions that are best studied using family analysis.
Before running an application workflow, it is important to prepare the sequencing reads as explained in the following diagram (figure 19.1).
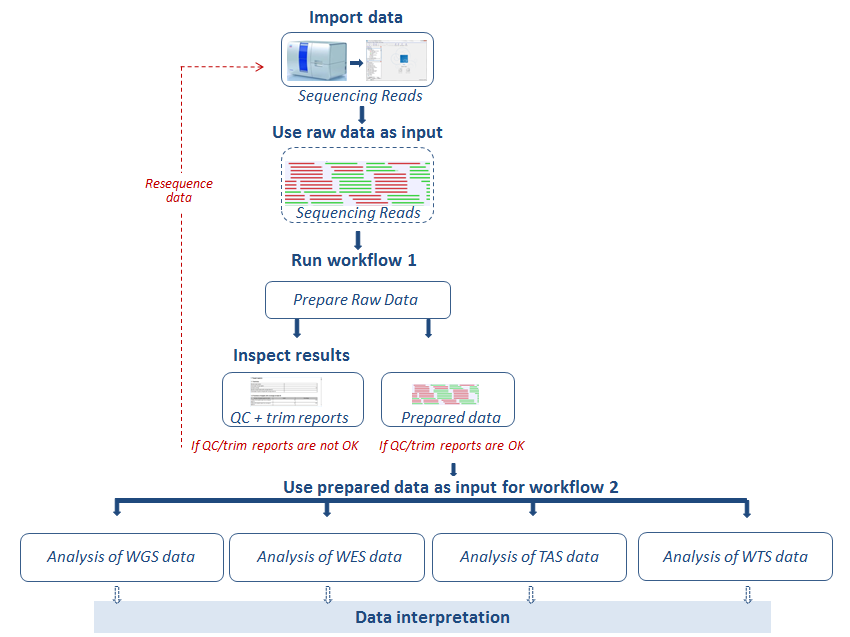
Figure 19.1: From sequencing reads to data interpretation.
The workflows are specific to the type of data used as input: Whole Genome Sequencing (WGS), Whole Exome Sequencing (WES), Targeted Amplicon Sequencing (TAS) and Whole Transcriptome Sequencing (WTS). For each of the first three categories, WGS, WES, and TAS, General Analysis workflows can be used for general identification and annotation of variants irrespective of disease. In Somatic Cancer you can find workflows designed specifically for cancer research. Finally, use Hereditary Disease workflows to study variants that cause rare diseases or hereditary diseases (HD).
The template workflows found under each of the first three applications have similar names (with the only difference that "WGS", "WES", or "TAS" have been added after the name). However, each have been tailored to the individual applications with parameter settings that have been adjusted to fit the expected differences in coverage between the different application types. We therefore recommend that you use the template workflow that is found under the relevant application heading.
Subsections
