Generate MSI Baseline
The tool Generate MSI Baseline can be used to generate microsatellite instability (MSI) baseline tracks. The tool can be found in the Toolbox:
Toolbox | Biomedical Genomics Analysis (![]() ) | Oncology Score Estimation (
) | Oncology Score Estimation (![]() ) | Generate MSI Baseline (
) | Generate MSI Baseline (![]() )
)
To generate a baseline, select at least five read mappings from microsatellite stable (MSS) samples as input. For better results, we recommend using 15 samples or more.
In the next step, the following parameters can be adjusted (figure 8.7):
- MSI loci track: The microsatellite loci track. It could be an existing MSI baseline.
- Flanking signature length: The length of the microsatellite locus in a read is determined by identifying flanking signatures on each side of the locus. The flanking signature should be long enough to be unique in the read, while short enough to be present in as many reads as possible. By default, the length of the flanking signature is 8 bp.
- Allow one mismatch in the flanking signature: When enabled, the flanking signatures are accepted even if it has one SNP compared to the reference sequence.
- Ignore broken pairs: When enabled, broken pairs in paired end reads are ignored for calculating the microsatellite length distribution.
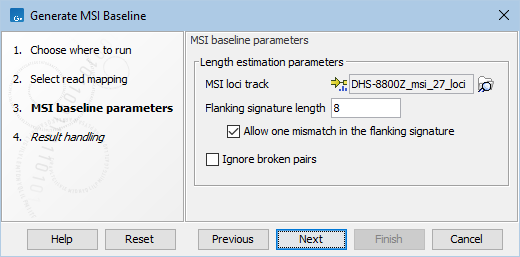
Figure 8.7: Parameters for Generate MSI Baseline.
Note that all the MSI baseline tracks available in the QIAseq TMB Panels hg38 Reference Data Set have been generated from samples that were mapped against the hg38 (no alternative analysis set) reference sequence.
