Identify QIAseq xHYB Somatic Variants
The Identify QIAseq xHYB Somatic Variants template workflow calls somatic variants. It is suitable for data generated using QIAseq xHYB panels without addition of mitochondrial spike-in probes. If mitochondrial spike-in probes have been added, the Identify QIAseq xHYB Somatic Variants including Mitochondrial should be used (see Identify QIAseq xHYB Somatic Variants including Mitochondrial).
It can be found at:
Template Workflows | Biomedical Workflows (![]() ) | QIAseq Sample Analysis (
) | QIAseq Sample Analysis (![]() ) | QIAseq DNA workflows (
) | QIAseq DNA workflows (![]() ) | Identify QIAseq xHYB Somatic Variants (
) | Identify QIAseq xHYB Somatic Variants (![]() )
)
This workflow can also be launched from the Analyze QIAseq Samples guide, which is described in The Analyze QIAseq Samples guide. It is available under the xHYB Human tab.
If you are connected to a CLC Server via your Workbench, you will be asked where you would like to run the analysis. We recommend that you run the analysis on a CLC Server when possible.
The initial steps of the workflow for selecting reads and the appropriate reference data, as well as mapping reads to the reference, are similar to the Identify QIAseq Exome Germline Variants workflow, which is described in Identify QIAseq Exome Germline Variants.
For variant detection, the Low Frequency Variant Detection tool is used. The parameters for this tool can be set in the relevant wizard step when launching the workflow (figure 14.59). For a description of the different parameters that can be adjusted, see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Low_Frequency_Variant_Detection.html. If you click on "Locked Settings", you will be able to see all parameters used for variant detection in the template workflow (figure 14.59).
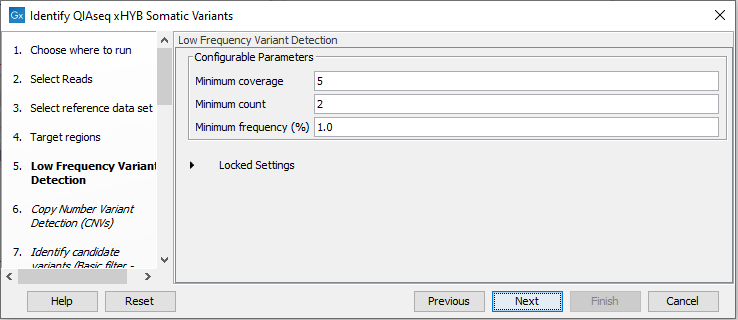
Figure 14.59: Specify the parameters for the Low Frequency Variant Detection tool.
The Copy Number Variant Detection tool is used for detection of copy number variants when control mappings are provided, as described for the Identify QIAseq Exome Germline Variants workflow in Identify QIAseq Exome Germline Variants. Mapping of control samples can be performed using the workflow described in Create QIAseq Exome CNV Control Mapping and specifying the relevant xHYB target regions in the Target regions wizard step.
The settings of a comprehensive variant filtering cascade can be adjusted in a series of wizard steps.
In the final wizard step, choose to Save the results of the workflow and specify a location in the Navigation Area before clicking Finish.
Subsections
