Appendix A - Sequence file naming pattern
RNA-seq Analysis Portalsupports analysis of FASTQ files. To be appropriately grouped into samples during upload, the sequence files names must adhere to the standard Illumina file naming. If you have FASTQ files from e.g. a Thermo Fisher Ion Torrent sequencer, you can manually rename them to fit your grouping needs:
<sample-name>_S<sample_number>_L<lane (0-padded to 3 digits)>_R<read number>_001.fastq(.gz)
Example of single lane, paired end data set consisting of two FASTQ files that will be grouped into one sample, TC-35-A_S1:
TC-35-A_S1_L001_R1_001.fastq.gz TC-35-A_S1_L001_R2_001.fastq.gz
Example of a four lane, paired end data set consisting of eight FASTQ file that will be grouped into one sample, 501-708-SEQC-77-1_S1:
501-708-SEQC-77-1_S1_L001_R1_001.fastq.gz 501-708-SEQC-77-1_S1_L001_R2_001.fastq.gz 501-708-SEQC-77-1_S1_L002_R1_001.fastq.gz 501-708-SEQC-77-1_S1_L002_R2_001.fastq.gz 501-708-SEQC-77-1_S1_L003_R1_001.fastq.gz 501-708-SEQC-77-1_S1_L003_R2_001.fastq.gz 501-708-SEQC-77-1_S1_L004_R1_001.fastq.gz 501-708-SEQC-77-1_S1_L004_R2_001.fastq.gz
For single lane, single read data, as what may you see from the QIAseq miRNA Library Kit, each sample is made up from one FASTQ file. The following two FASTQ files are converted into the two samples, QL4_S4 and QL6_S6.
QL4_S4_L001_R1_001.fastq QL6_S6_L001_R1_001.fastq
On the RNA-seq Analysis Portal sequencing data uploader tab, once you have selected your FASTQ files, the Files to samples section on the right provides a sample grouping preview (figure 59). If grouping is not as expected this is likely due to the FASTQ file names not following the expected format.
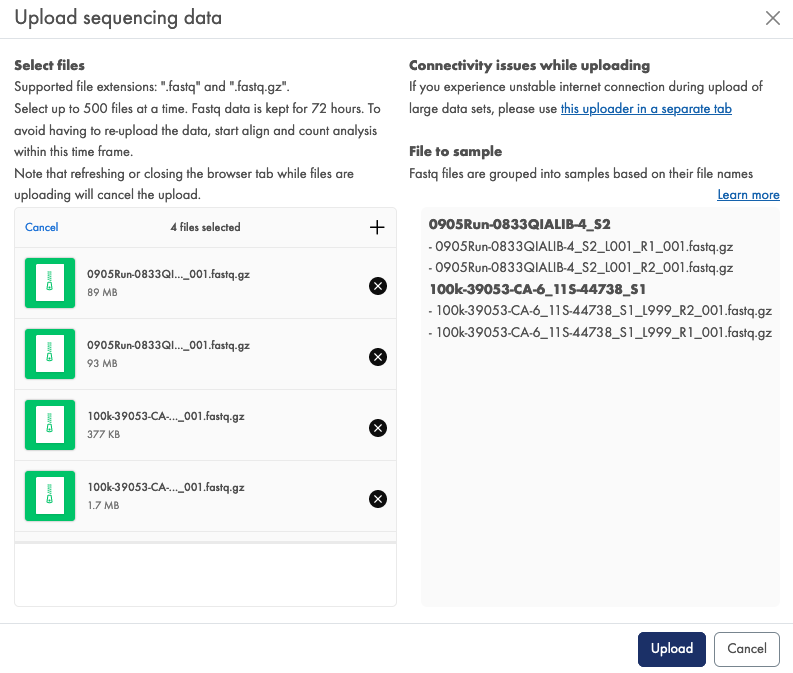
Figure 59: The Files to samples preview provides a check of sample grouping.
