Find Resistance with PointFinder
The Find Resistance with PointFinder tool finds known antimicrobial resistance conferring mutations in NGS reads (from isolates or shotgun metagenome samples after binning) and summarizes them in a concise and easily understandable report. In contrast to the Find Resistance with Nucleotide DB tool that quantifies the occurrence of entire resistance conferring genes, the aim here is to detect the presence of resistance conferring mutations in antibiotic targets in both susceptible and resistant strains.
The presence of antibiotic resistance conferring variants is inferred by running the read mapper against a database containing both wild-type and known resistant mutants of antibiotic target genes. The source for underlying information about resistance conferring mutations is based on the web tool PointFinder data [Zankari et al., 2017], with added information from other resources. This database can be downloaded using the Download Resistance Database tool, where you currently have the choice to download known variants in 5 common pathogens (figure 14.1).

Figure 14.1: Known variants in these common pathogens can be downloaded with the Download Resistance Database.
To run Find Resistance with PointFinder, go to:
Toolbox | Microbial Genomics Module (![]() ) | Drug Resistance Analysis (
) | Drug Resistance Analysis (![]() ) | Find Resistance with PointFinder (
) | Find Resistance with PointFinder (![]() )
)
The tool accepts a nucleotide sequence or sequence list as input, followed by the selection of the PointFinder Database for the relevant pathogen.
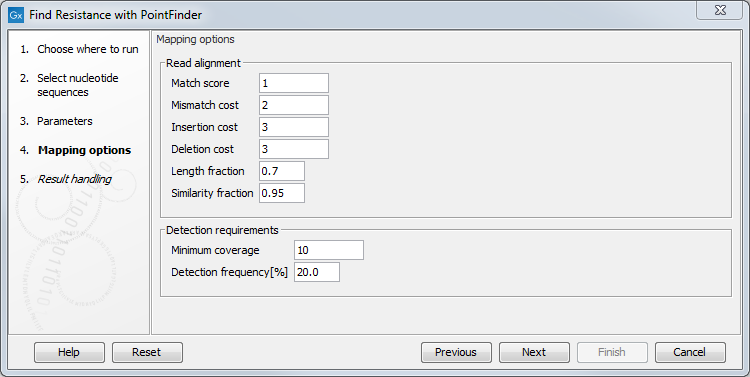
For the read mapping step itself, several parameters are available (figure 14.2):
- Match score: score for a match
- Mismatch cost: cost for a mismatch
- Insertion cost: cost for an insertion
- Deletion cost: cost for accepting a deletion
- Length fraction: minimum length of the mapped portion of the read
- Similarity fraction: minimal fraction of matches in the mapped region
The tool first maps the reads to the specified database sequences. Next, the tool analyses the read mappings. For each reference sequence containing the variant, the tool verifies if the reads mapping to that reference or related references (e.g. references with an additional mutation) contain that variant. The result table will only list those entries from the database that have a minimum coverage and exceed the minimum frequency threshold. The tool also adds information:
- Minimum coverage: the number of reads covering the variant region. The coverage must be complete, e.g. all 3 nucleotides that constitute an amino acid change have to be covered.
- Detection frequency: The minimum allele frequency that is required to annotate a variant as being present in the sample.
The tool outputs a table containing information about the variants detected in the reads. The columns available in that table can be seen in figure 14.3 (which also shows an example of report output by the tool):
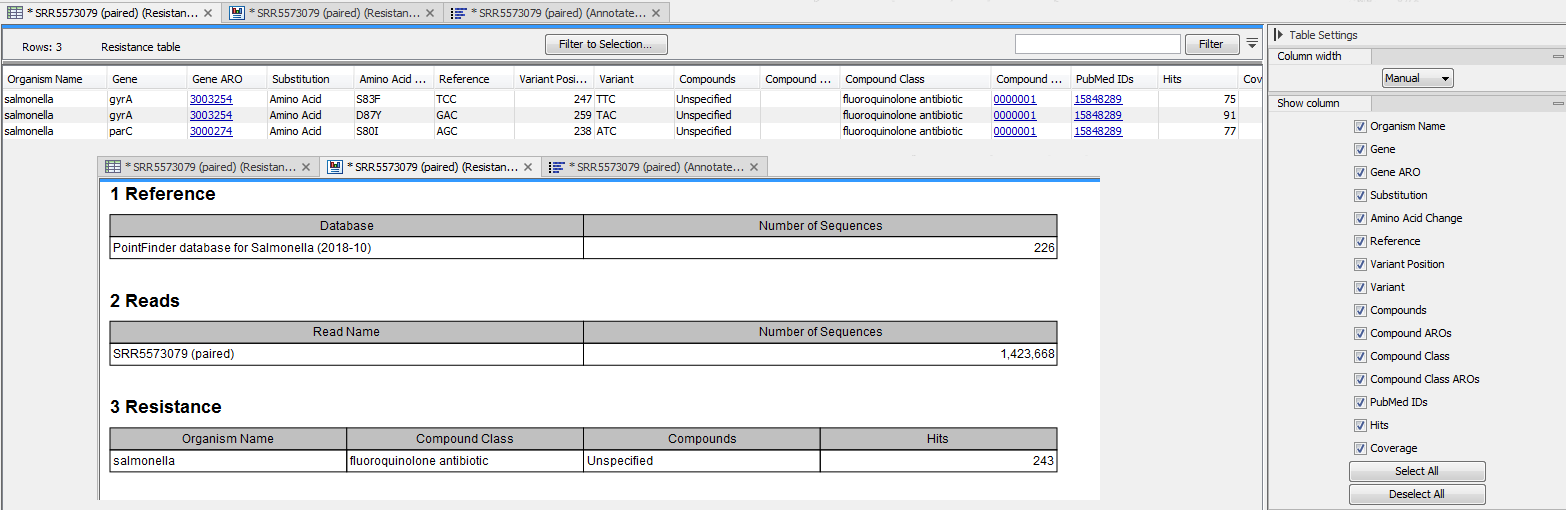
Figure 14.3: Table generated by Find Resistance with PointFinder, shown here together with the corresponding report.
By enabling the "Output annotated reads" option, the user can obtain a copy of the subset of reads that map to target variants. The reads will be annotated with the following annotations:
- Target: The name of the target reference (from the PointFinder database) where the read mapped to, e.g. "salmonella_gyrA_AAS_S_83_Y_TCC_247_TAC".
- Compound Class: The compound class to which the variant gives resistance, e.g. fluoroquinolone antibiotic.
- Compounds: The compound(s) class to which the variant gives resistance, e.g. colistin.
The resulting report contains information about the reads and the database that was used.