Create Taxonomic Profiling Index
This tool will compute a a taxonomic profiling index from a reference database. Indexes are then used as input to the Taxonomic Profiling tool. The computation of index files for taxonomic profiling is memory and hard-disk intensive due to the large sizes of reference databases usually employed for this task. The algorithm requires roughly the number of bases in bytes of memory (as indicated by the Download Custom Microbial Reference Database tool), i.e., approximatively the size of the uncompressed reference database; and twice this amount in hard disk space.
To run the tool, go to:
Toolbox | Microbial Genomics Module (![]() ) | Databases (
) | Databases (![]() ) | Taxonomic Analysis | Create Taxonomic Profiling Index (
) | Taxonomic Analysis | Create Taxonomic Profiling Index (![]() )
)
In the first dialog, select one or several nucleotide databases, for example downloaded with the Download Custom Microbial Reference Database tool or the prokaryotic databases from the Download Curated Microbial Reference Database tool (figure 17.8). Note that sequences with identical Assembly ID annotations are treated as the same reference, see Using the Assembly ID annotation.
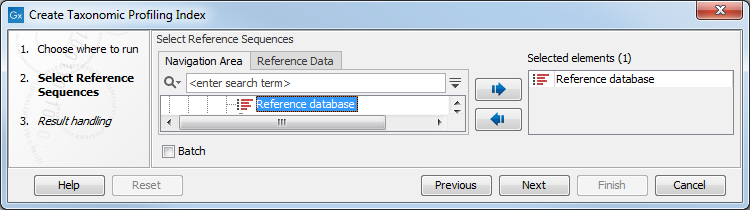
Figure 17.8: Select reference sequences.
The output is an index file and a report as seen in figure 17.9. The report list the number of sequence and basepairs that were indexed.
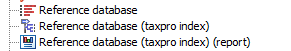
Figure 17.9: The reference sequences,index and report as seen in the Navigation Area.
