Download Pathogen Reference Database
Download a collection of bacterial assemblies and enrich with metadata from the NCBI Pathogen Detection Project (see http://www.ncbi.nlm.nih.gov/projects/pathogens/).
Toolbox | Microbial Genomics Module (![]() ) | Databases (
) | Databases (![]() ) | Taxonomic Analyses (
) | Taxonomic Analyses (![]() ) | Download Pathogen Reference Database (
) | Download Pathogen Reference Database (![]() )
)
This will open the following wizard window (figure 17.7):
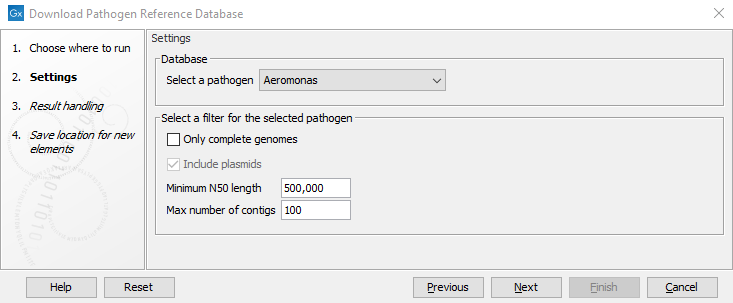
Figure 17.7: Downloading assemblies and metadata for a selected pathogen from the NCBI Pathogen Detection Project.
The settings are:
- Select a pathogen. Select a pathogen for which to download assemblies and associated metadata.
- Only complete genomes. This can be used to switch between complete genomes or to also allow for downloading incomplete assemblies.
- Include plasmids. This option can be used to include or exclude plasmids from the downloaded database. Note that if a database of plasmids only is required, the Download Custom Microbial Reference Database tool should be used instead.
- Minimum N50 length. This option can be used to remove assemblies with shorter N50 values (the default value is set at 500,000 bp). Short N50 values typically indicate low assembly quality. This option is not available when "Only complete genomes" has been selected.
- Maximum number of contigs. This option can be used to remove assemblies with a higher number of contigs (the default value is set at 100). Many contigs typically indicate low assembly quality. This option is not available when "Only complete genomes" has been selected,
Specify a location to save the database. We recommend to create a folder where you can save all the databases and MLST schemes necessary to run some of the CLC Microbial Genomics Module tools.
The resulting database includes a list of different bacterial genome sequences as well as the associated accession numbers, descriptions, taxonomy and size of the sequences. In addition, each reference genome will be annotated with the following metadata (when available):
- serovar
- strain
- taxonomy
- sample collection date
- geographical location
- isolation source
- host
- host disease
- outbreak
- SRA run id
- SRA project id
Once a database has been downloaded, it is possible to extract a subset following the instructions described in Extracting a subset of a database.
