Create DIAMOND Index
This tool will compute a DIAMOND index from a protein database. These indexes can then be used as input to the Annotate CDS with Best DIAMOND Hit tool (see section Annotate CDS with Best DIAMOND Hit) and Annotate with DIAMOND tool (see section Annotate with DIAMOND).
If the input sequences contain metadata, such as GO-terms, these will be transferred to the created index.
To run the tool, go to:
Toolbox | Microbial Genomics Module (![]() ) | Databases (
) | Databases (![]() ) | Functional Analysis (
) | Functional Analysis (![]() ) | Create DIAMOND Index (
) | Create DIAMOND Index (![]() )
)
In the first dialog, select a protein database downloaded with the Download Protein Database tool (figure 18.6).
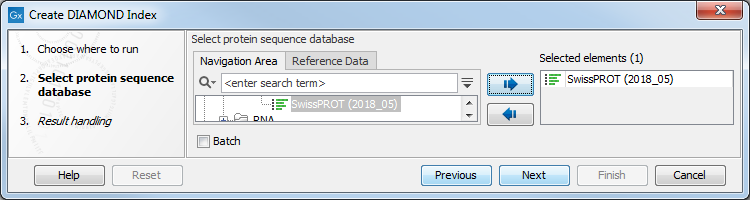
Figure 18.6: Select a protein database.
The output is an index file as seen in figure 18.7.
![]()
Figure 18.7: The protein database and its index as seen in the Navigation Area.
The default view for the DIAMOND index is a tabular overview of the sequence entries and their associated metadata, such as GO-terms (see figure 18.8).
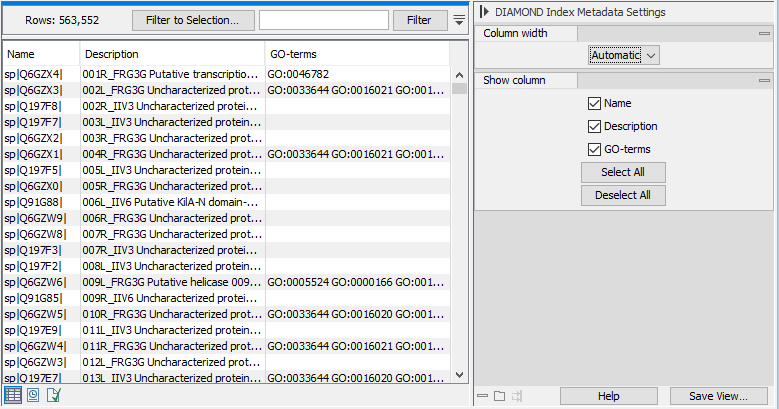
Figure 18.8: The default view for the DIAMOND index element.
