Database Builder
If you left Skip Database Builder unchecked, assembly data is not downloaded right away. Instead, the Database Builder will open (figure 17.5).The Database Builder table contains a range of metadata. This takes a bit to download, why opening the table may take a little while. The metadata is based on information from GenBank, http:///www.ncbi.nlm.nih.gov/genbank/.html.
With the Database Builder you can further customize the reference set to be downloaded:
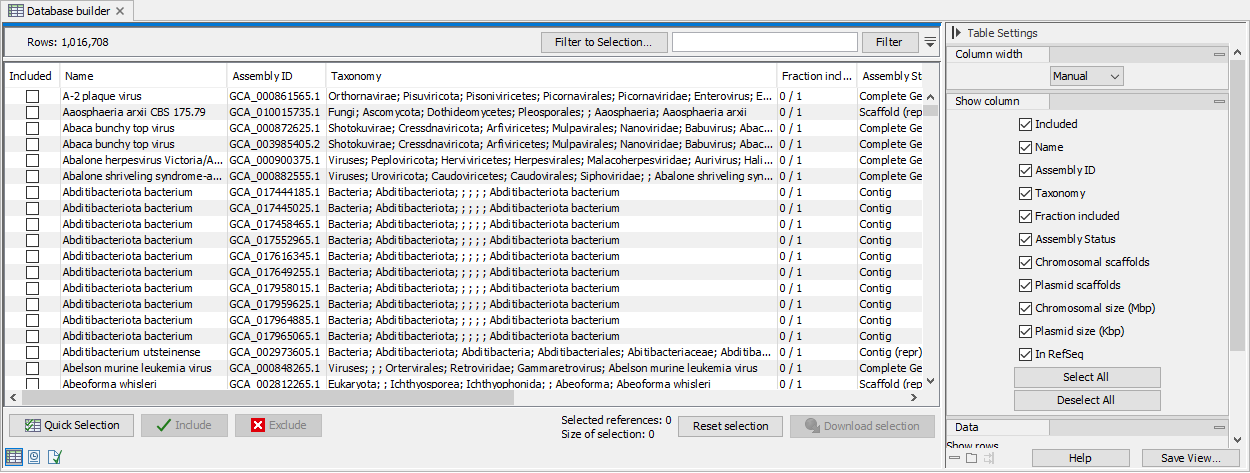
Figure 17.5: Search, filter and select assemblies to download
- Use the Quick selection button below the table to immediately download one of the following predefined subsets:
- Single scaffold complete genomes in RefSeq
- Complete genomes in RefSeq
- All complete genomes
- Aggregate the table to a specified taxonomic group using the drop down menu in the "Data" palette of the side panel. Use the category "Name" to de-aggregate the table.
- Use filter(s) and select row by dragging or pressing Ctrl+A to keep only the rows you are interested in, and click on the button Include to stage the selected references for downloading, which is indicated by a checkmark in the "Included" column.
- Alternatively, press Ctrl+A then click on Include to include all rows first. Then set one or several filters, and use the button Exclude on the remaining rows. Clear the filter(s) by clicking on the red buttons next to each filter set. The rows not filtered away in the second step should still be checked.
- Use the Reset button to reset the selection. This restores the builder to the initial state with only pre-selected reference included. If no references were pre-selected this will exclude all.
Once the table has all the desired references included, which is indicated by a checkmark, click Download selection. Close to the button, you can check how many references are selected and see an estimate of the total size of the selection.
The dialog shown in figure 17.6 allows you to include all annotation tracks (annotation tracks are not needed for taxonomic profiling applications, but required when creating MLST schemes). An additional filter, Minimum contig length, may also be specified (this option is not available when downloading the curated database). It also warns about the memory and disk requirements that will be needed to later run the Taxonomic Profiling tool with the database you are about to download.
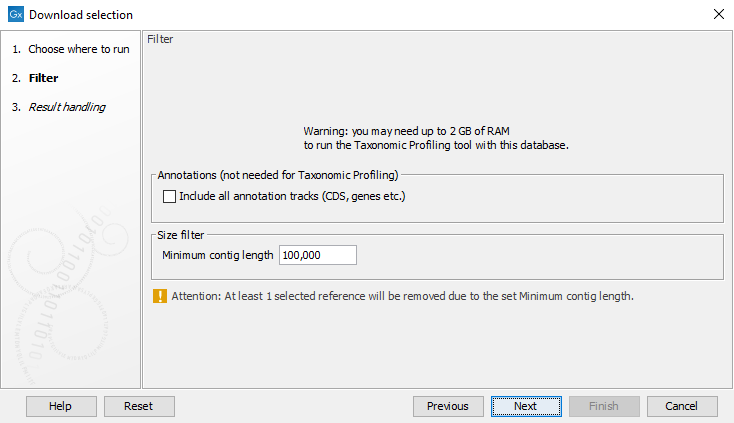
Figure 17.6: Filter options for download of the selected references
Once a database has been downloaded, it is possible to extract a subset following the instructions described in 17.1.1
