Assign Taxonomies to Sequences in Abundance Table
The Assign Taxonomies to Sequences in Abundance Table tool lets you add taxonomies to abundance table features that have sequences associated. This is useful for annotating amplicon sequence variant (ASV) tables, and OTU tables with de novo OTUs where sequences are not annotated by the initial analysis tools.
The tool requires a reference index and works by mapping each sequence from the abundance table to this reference index. The underlying analysis is the same as for Taxonomic Profiling, see Taxonomic Profiling.
Creating the required reference index
You create a reference index using Create Taxonomic Profiling Index, see Create Taxonomic Profiling Index. As input, you will need a reference database, i.e. a sequence list containing reference sequences with taxonomy annotations.
Reference database can be obtained using one of the Download Database tools. The choice of reference database depends on your data.
For amplicon data, consider the reference databases available with Download Amplicon-Based Reference Database, see Download Amplicon-Based Reference Database.
For whole genome data, you may use the databases from Download Curated Microbial Reference Database, see Download Curated Microbial Reference Database. Alternatively, create your own reference database with Download Custom Microbial Reference Database, see Download Custom Microbial Reference Database.
Running the tool
To run the Assign Taxonomies to Sequences in Abundance Table tool:
Toolbox | Microbial Genomics Module (![]() ) | Metagenomics (
) | Metagenomics (![]() ) | Abundance Analysis (
) | Abundance Analysis (![]() ) | Assign Taxonomies to Sequences in Abundance Table (
) | Assign Taxonomies to Sequences in Abundance Table (![]() )
)
Select the abundance table with sequences to be annotated.
Select the reference index to map the abundance table sequences to (figure 7.1).
Choose settings for taxonomic assignment:
- Minimum similarity percentage Sequences in the abundance table must be at least this similar to sequence in the reference index to be matched and get a new taxonomy assigned.
- Clear existing taxonomy All existing abundance table taxonomy annotations are removed. Only abundance table sequences with a reference index match will get a taxonomy assignment.
- Overwrite existing taxonomy Abundance table sequences with a reference index match will get a new taxonomy assigned. Sequences with no match will retain the existing taxonomy annotation.
- Use existing taxonomy when present Only abundance table sequences that do not already have a taxonomic annotation will get a new taxonomy assigned.
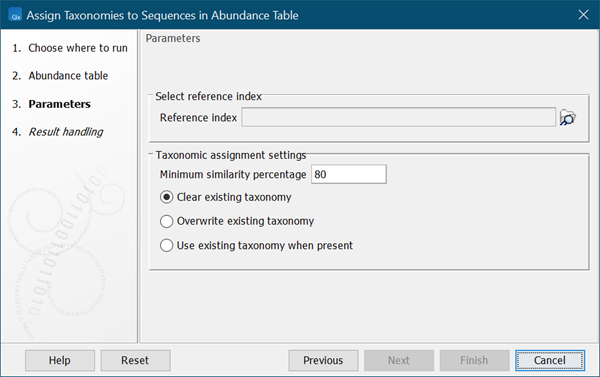
Figure 7.1:
Select reference index and set parameters for taxonomic assignment.
Select Create Report to generate a report with summary information on taxonomic assignment.
The output
- Abundance table with taxonomy annotations The new taxonomy assignments are listed in the Taxonomy column.
- Assign taxonomies report The report contains the following sections:
- Summary Information on the sequences in the abundance table.
- Reference index summary Information on the reference index.
- Taxonomy assignment
- Sequences with reference index match: Sequences that met the Minimum similarity treshold.
- Taxonomy assigned (was blank): Taxonomy was blank, new taxonomy has been assigned.
- Taxonomy updated: The existing taxonomy has been replaced.
- Existing taxonomy retained: The existing taxonomy is retained. This can happen when the taxonomy of the matched reference index sequence is identical to the existing taxonomy, or when Taxonomy assignment was set to Use existing taxonomy when present.
- Sequences with no reference index match (insufficient similarity): Sequences that did not meet the Minimum similarity threshold.
- Existing taxonomy retained: Sequences for which an existing taxonomy remains.
- No taxonomy: Sequences with no taxonomy.
- Sequences with reference index match: Sequences that met the Minimum similarity treshold.
