Build Functional Profile
To compute the number of reads in a sample mapping to regions involved with Pfam domains, or BLAST or DIAMOND hits, you can run the Build Functional Profile tool by going to:
Toolbox | Microbial Genomics Module (![]() ) | Functional Analysis (
) | Functional Analysis (![]() ) | Build Functional Profile (
) | Build Functional Profile (![]() )
)
In the first wizard (figure 13.11), select the read mapping for which you want to build the functional profile.
Figure 13.11: Select a read mapping.
The parameters that can be set are seen in figure 13.12:
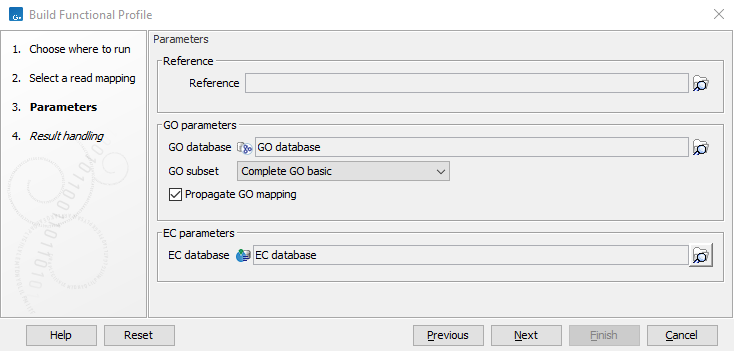
Figure 13.12: Specify a reference, a GO database and an EC database.
- Reference. A reference set of contigs annotated with Pfam domains, BLAST, or DIAMOND hits. If the read mapping contains an annotated genome, this parameter is optional.
- GO database. The GO database. If the reference contains Pfam domains, this database can be used to map from Pfam domains to GO terms. If the BLAST or DIAMOND hits contain GO-terms annotations, this will be also matched against the database and appear in the GO abundance table output. The GO database can be downloaded using the Download Ontology Database tool (see section "Download GO database" ).
- GO subset. A subset of the GO database. Since many GO terms are too general or too specific, several meaningful subsets of GO terms are provided. See http://geneontology.org/docs/download-ontology/.
- Propagate GO mapping. When selected, GO terms are mapped to all their ancestor terms. For example, the Pfam domain "CutC" maps to the GO term "0005507 // copper ion binding". If Propagate GO mapping is enabled, the tool would also map to more general GO terms such as "0055070 // copper ion homeostasis", "0055076 // transition metal ion homeostasis", and "0065007 // biological regulation".
- EC database. The EC database. If the reference contains BLAST or DIAMOND domains, this database can be used to map EC terms. The EC database can be downloaded using the Download Ontology Database (see section "Download GO database" ).
You can then select which output elements should be generated figure 13.13.
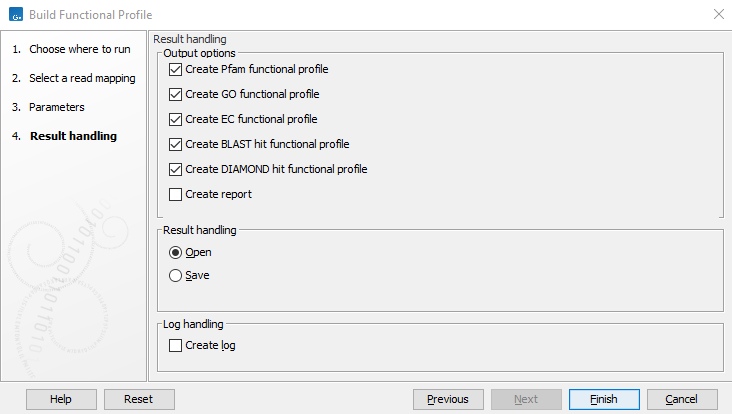
Figure 13.13: Specify what type of output you want the tool to generate.
- Create Pfam functional profile. Abundance table obtained by counting reads overlapping Pfam domains.
- Create GO functional profile. Abundance table obtained by counting reads overlapping GO terms. Note that a database must be specified in order to build a GO functional profile. For BLAST and DIAMOND hits, the GO-terms specified on the annotations are used, but preexisting GO annotations on pfam domains are ignored by this tool.
- Create EC functional profile. Abundance table obtained by counting reads overlapping EC terms. Note that a database must be specified in order to build an EC functional profile.
- Create BLAST hit functional profile. Abundance table obtained by counting reads overlapping BLAST hits.
- Create DIAMOND hit functional profile. Abundance table obtained by counting reads overlapping DIAMOND hits.
- Create Report. A report stating statistics about the input reference contigs and read mapping, as well as the number of matches to each feature.
The resulting functional abundance tables store the number of reads corresponding to each Pfam domain, GO term, EC number, best BLAST hit or best DIAMOND hit.
Subsections
