Download Resistance Database
The Download Resistance Database tool can download databases for the Find Resistance with Nucleotide DB, Find Resistance with PointFinder and Find Resistance with ShortBRED tools.
To run the tool, go to:
Toolbox | Microbial Genomics Module (![]() ) | Databases (
) | Databases (![]() ) | Drug Resistance Analysis (
) | Drug Resistance Analysis (![]() ) | Download Resistance Database (
) | Download Resistance Database (![]() )
)
The following databases are available (figure 19.1):
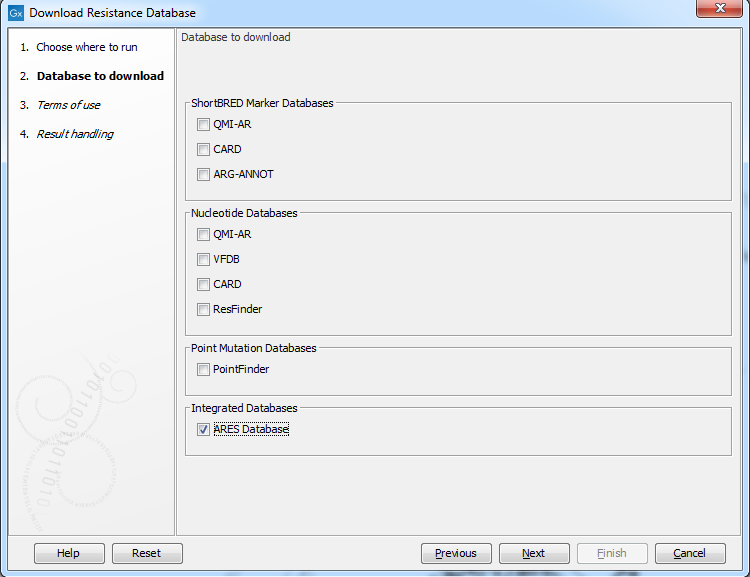
Figure 19.1: Known variants in these common pathogens can be downloaded with the Download Resistance Database.
ShortBRED Marker Databases
These databases can be used with the Find Resistance with ShortBRED tool. The databases are marker databases, containing peptide fragments that uniquely characterize sets of similar proteins, rather than a specific gene.
- QMI-AR The QIAGEN Microbial Insight - Antimicrobial Resistance database is a curated database containing peptide markers derived from the following source databases: CARD (https://card.mcmaster.ca/), ARG-ANNOT (http://backup.mediterranee-infection.com/article.php?laref=282&titre=arg-annot), NCBI Bacterial Antimicrobial Resistance Reference Gene Database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA313047), ResFinder (https://bitbucket.org/genomicepidemiology/resfinder_db/src/master/).
- CARD This database contains peptide markers derived from the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/).
- ARG-ANNOT This database contains a set of protein marker sequences based on the ARG-ANNOT database.
Nucleotide Databases
These databases can be used with the Find Resistance with Nucleotide DB tool. The databases contain full nucleotide gene sequences.
- QMI-AR The QIAGEN Microbial Insight - Antimicrobial Resistance database is a curated database containing nucleotide sequences compiled from the following source databases: CARD (https://card.mcmaster.ca/), ARG-ANNOT (http://backup.mediterranee-infection.com/article.php?laref=282&titre=arg-annot), NCBI Bacterial Antimicrobial Resistance Reference Gene Database (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA313047), ResFinder (https://bitbucket.org/genomicepidemiology/resfinder_db/src/master/).
- VFDB This database contains nucleotide sequences compiled from the Virulence Factor database, core dataset (http://www.mgc.ac.cn/VFs/download.htm).
- CARD This database contains nucleotide sequences compiled from the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/).
- ResFinder (DNA) The output is a sequence list with modifiable metadata relating the sequences to drug resistance information (see https://cge.cbs.dtu.dk/services/data.php). In the table view, users can review and customize the metadata associated with the sequences.
Point Mutation Databases
These databases are used with the Find Resistance with PointFinder tool. The databases contain information about mutations in genes.
- PointFinder This database contains wild type genes and known resistance conferring mutations for a number of organisms. The database source can be found here: https://bitbucket.org/genomicepidemiology/.
Integrated Databases
- ARES Database The ARES Database is an interactive interface to ARESdb: https://www.sciencedirect.com/science/article/pii/S1672022919300920, a curated database of computationally inferred antimicrobial resistance genes and their respective predictive performances, both established and provided by Ares Genetics. With the ARES Database it is possible to produce sequence lists which are compatible with the Find Resistance with Nucleotide DB and Find Resistance with PointFinder tools, but also with other tools taking annotated sequence lists as input.
Subsections
