The report output from Single Cell V(D)J-Seq Analysis
The optional report includes information for different chain types:
- For TCR Cell Clonotypes: TRA + TRB, TRA, TRB, TRG + TRD, TRG, and TRD;
- For BCR Cell Clonotypes: IGH + IGK, IGH + IGL, IGH, IGK, and IGL.
|
Ideally, for each barcode all chains have been identified that together form the complete receptor: both TRA + TRB or TRG + TRD chains for T cell, and both the two heavy (IGH) and two light (IGK or IGL) chains for B cells. For B cells, it is not possible to know which of the heavy chains is connected to which of the light chains. Hence, when reporting information for IGH + IGK and IGH + IGL, barcodes can provide fractional counts. For example, if a barcode has three identified chains, IGH, IGH and IGK, then this barcode contributes one IGH + IGK clonotype. However, since it is unknown which of the two heavy chains is connected to the IGK chain, when reporting detailed information about IGH + IGK clonotypes, such as the CDR3 length, this barcode will contribute 1/2 for both possible combinations. |
Only the chain types that are found in the Cell Clonotypes are present in the report.
The following information is provided for each different chain type:
- Summary. Summary tables with information about the performed assembly, trimming and identified clonotypes. See The clonotype identification algorithm for more details.
- Diversity indices. Several diversity indices, as listed below. The extrapolated diversity gives a projection of what the diversity would have been if the sample had been sequenced deeply enough to identify all clonotypes.
- Distinct clonotypes: The number of different clonotypes detected.
- Extrapolated diversity (chaoE): The extrapolated number of detected distinct clonotypes as described in [Chao, 1987].
- Lorenz curve at 50% of total: The fraction of all detected clonotypes that account for 50% of the total count. Also sometimes denoted as D50.
- Inverse Simpson's index: Let
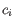 denote the count for the
denote the count for the 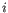 th distinct clonotype and let
th distinct clonotype and let
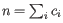 . Then the inverse Simpon's index is defined as:
. Then the inverse Simpon's index is defined as:
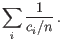
- Extrapolated Inverse Simpson's index (chaoE): The extrapolated inverse Simpson's index as described in [Chao et al., 2014].
- Shannon-Wiener index: With
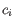 and
and 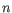 defined as above, the Shannon-Wiener index is defined as:
defined as above, the Shannon-Wiener index is defined as:
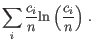
- Extrapolated Shannon-Wiener index (chaoE): The extrapolated Shannon-Wiener index as described in [Chao et al., 2013].
- Rarefaction. Rarefaction curves, also known as species accumulation curves. They show the expected number of distinct clonotypes discovered as a function of the total number of detected clonotypes, together with the confidence interval (CI), obtained from a normal approximation. The curve is
- interpolated down to 0 clonotypes;
- extrapolated to twice the total number of detected clonotypes.
- CDR3 length. The distribution of the length of the CDR3 nucleotide sequences for all detected clonotypes. Peaks are expected every 3 nucleotides due to repertoires consisting predominantly of in-frame CDR3 sequences.
- V, D, J and C usage. Bar plots showing the V, D, J and C segment usage for all detected clonotypes.
- Frequencies. The percentage of all detected clonotypes that are unique and the clonotype abundance: how many distinct clonotypes are found with abundance (count)
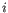 . Most clonotypes are expected to be unique, so the percentage is close to 100% and most clonotypes have abundance 1.
. Most clonotypes are expected to be unique, so the percentage is close to 100% and most clonotypes have abundance 1.
- Productive summary. The percentage of all detected clonotypes that have productive CDR3 nucleotide sequences, and the percentage of barcodes with at least one productive CDR3 nucleotide sequence.
Note that for diversity indices and rarefaction, the number of distinct clonotypes is used. For the rest of the report, the number of detected clonotypes contains all clonotypes for all barcodes, where if more than one barcode has the same clonotype, this is counted multiple times.
