Immune Repertoire
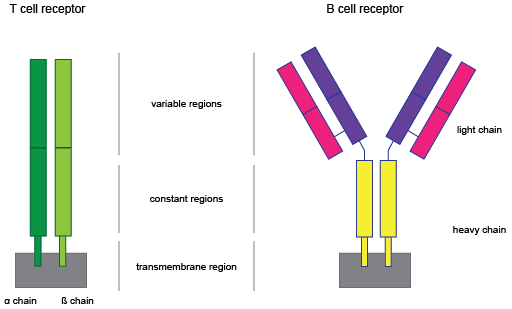
Figure 10.1: T and B cell receptors structure.
T and B cells form our acquired immune response. They both contain highly variable receptors (see figure 10.1) that recognize foreign antigens and consist of multiple chains. T cell receptors (TCR) consist of two chains, either an ![]() (TRA) and a
(TRA) and a ![]() (TRB) chain, or a
(TRB) chain, or a ![]() (TRG) and a
(TRG) and a ![]() (TRD) chain. B cell receptors (BCR) contain two light chains, and two heavy chains. There are two types of light chains in humans:
(TRD) chain. B cell receptors (BCR) contain two light chains, and two heavy chains. There are two types of light chains in humans: ![]() (IGK) and
(IGK) and ![]() (IGL), while other animals also contain other types of light chains. Once set, the light chain class remains fixed for the life of the B cell. There are five types of heavy chains (IGH) for mammals:
(IGL), while other animals also contain other types of light chains. Once set, the light chain class remains fixed for the life of the B cell. There are five types of heavy chains (IGH) for mammals: ![]() ,
, ![]() ,
, ![]() ,
, ![]() and
and ![]() , defining the class of the receptor.
, defining the class of the receptor.
The chains are encoded by genes that undergo somatic recombination. During this process, gene-segments are joined with random nucleotides at the junction sites. There are two types of recombination (see figure 10.2):
- VJ recombination, where one V (variable) gene-segment is joined to a J (joining) gene-segment;
- VDJ recombination, where a D (diversity) gene-segment is added between the V and J gene-segments.
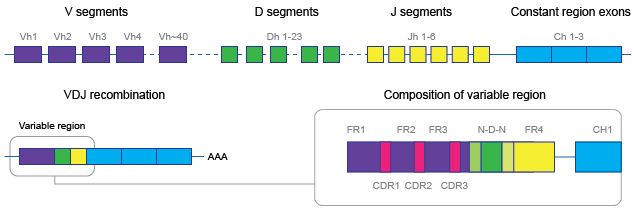
Figure 10.2: VDJ recombination brings together a V, D, J and C gene-segment.
For both types of recombination, a C (constant) gene-segment is also added following the J segment.
The TRA and TRG chains are the result of VJ recombination, while the TRB and TRD chains are the result of VDJ recombination. The V, D, J and C gene-segments are specific for each TCR chain type.
BCR light chains are the result of VJ recombination, while BCR heavy chains are the result of VDJ recombination. BCR heavy chains have three to four C gene-segments. The V, D, J and C gene-segments are specific for each BCR light chain type, while they are shared by the BCR heavy chains.
The V and J segments contain a conserved cysteine (C) and phenylalanine/tryptophan (F/W) amino-acid, marking the beginning and end of the CDR3 (complementary-determining) region, respectively. Due to inclusion of random nucleotides at the junctions between segments, the CDR3 region is highly variable. The V segment contains two other highly variable regions: CDR1 and CDR2 (see figure 10.2).
CLC Single Cell Analysis Module offers tools to clonotype reads and characterize the T or B cell receptor repertoire (Single Cell V(D)J-Seq Analysis), filter the repertoires (Filter Cell Clonotypes), combine them across samples (Combine Cell Clonotypes), compare them (Compare Cell Clonotypes) and convert them to cell annotations (Convert Clonotypes to Cell Annotations) for easy visualization on Dimensionality Reduction Plots.
Here, clonotyping consists of identifying which V, D, J and C segments from the reference data (Single Cell V(D)J-Seq Analysis) are used, and extracting the CDR3 region found between the conserved amino acids.
Subsections
- Single Cell V(D)J-Seq Analysis
- Filter Cell Clonotypes
- Combine Cell Clonotypes
- Compare Cell Clonotypes
- Convert Clonotypes to Cell Annotations
- The Cell Clonotypes element
