Export Peak Count Matrix
A peak count matrix can be exported to the following formats:
- Cell Ranger HDF5.
- MEX.
All options as shown in figure 3.7 are common to both exporters.
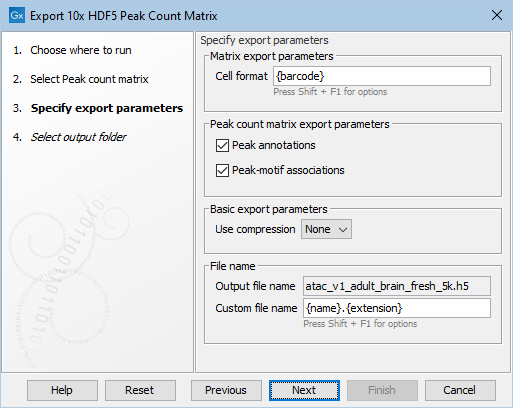
Figure 3.7: Options for export of peak count matrix.
- Cell format. Can be specified to include both the sample and barcode. Hover the mouse over this setting to see the possible options. Often the barcode in itself is sufficient as a unique identifier of cells. However, when an expression matrix contains multiple samples, the cell format must also include the sample, otherwise the export will fail.
- Peak annotations. If selected, the nearby genes will be exported to a separate file named peak_annotations.tsv.
- Peak-motif associations. If selected, the transcription factors will be exported to a separate file named peak_motif_mapping.bed.
The format of peak annotations and peak-motif annotations is as described in Import Peak Count Matrix (see Import Peak Count Matrix). The peak annotations have 6 columns.
- Use compression. Select among gzip, zip or no compression which is default.
- Output file name. Shows the name of the file. This can be customized by changing the default pattern in Custom file name.
