Import Expression Matrix
The following expression matrix formats can be imported into an Expression Matrix (
- Cell Ranger HDF5
- Loom
- MEX
- MEX archive
- CSV
Some other commonly encountered formats are specific to a programming language or software package. These can usually be exported from that software package as Loom files. For example:
- AnnData (h5ad). This format is defined by the AnnData package, and is used by Scanpy. It can be written to Loom using the `write_loom' method of the same package.
- .rds/.Robj. Data formats from the R programming language. These can often be written to Loom using the LoomR package, or methods in the same R package that was used to generate the files.
Options common to all importers
Several options are common to all expression matrix importers. Figure 2.1 shows the Cell Ranger HDF5 importer, which only contains these general options.
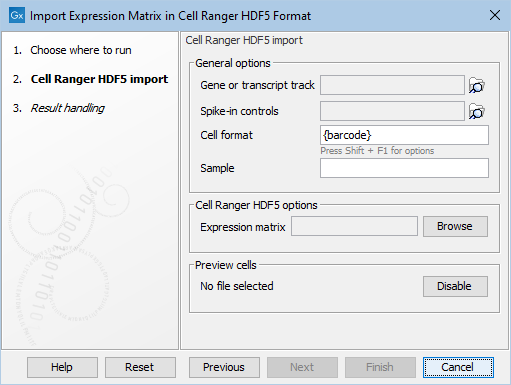
Figure 2.1: The Cell Ranger HDF5 importer. The General options are common to all the expression matrix importers.
- Gene or Transcript track. Genes or transcripts in the imported data are matched with features in the provided track to the extent possible. When a match is found, the genomic coordinates of the gene/transcript will be recovered. Matches are only found when the identification of the gene/transcript in the imported data with the feature in the track is unambiguous: one-to-many and many-to-one matches between the imported data and the provided track are not supported. This means, for example, that if a gene is present on two chromosomes of the track, then neither set of genomic coordinates will be recovered.
Matching is used to:
- View the Expression Matrix as a Track. For more information on tracks, see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Tracks.html.
- Define the mitochondrial chromosome when calculating the proportion of reads mapped to mitochondria in the QC for Single Cell tool (Count-based and extra-chromosomal filters).
- Recover identifiers (e.g. ENSG00000243485 for ENSEMBL genes) when these are not present in the input data. As identifiers are often more specific than e.g. gene names, this can help when training Cell Type Classifiers using the Train Cell Type Classifier tool (Train Cell Type Classifier), and when predicting cell types using a Cell Type Classifier.
The matching algorithm works by choosing an approach from the following list that maximizes the number of one-to-one matches between features in the provided track and features in the imported data:
- Matching names from the track with identifiers from the imported data
- Matching identifiers from the track with identifiers from the imported data
- Matching names from the track with unversioned identifiers from the imported data. An unversioned identifier is obtained by removing anything from or after the first '.' in the identifier. For example, ENSG00000243485 is the unversioned identifier for ENSG00000243485.5.
- Matching identifiers from the track with unversioned identifiers from the imported data
- Matching names from the track with names from the imported data
- Matching identifiers from the track with names from the imported data
In the case of a tie, the first equally good approach from the above list is used. If no matches are found, check that the correct Gene or Transcript track has been supplied.
- Spike-in controls. (Optional) Genes or transcripts in the imported data are also matched against the spike-in controls provided here. This is used when calculating the proportion of reads mapped to spike-in controls in the QC for Single Cell tool (Count-based and extra-chromosomal filters). It is also used to remove the spike-in controls from downstream analysis. For details on how to import spike-in controls, see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Import_RNA_spike_in_controls.html.
- Cell format and Sample. How cells are identified. See Cell format in importers for more details. When a file contains multiple samples, it is recommended to extract the sample name from the cell name. This allows the QC for Single Cell to process each sample separately, enables coloring of cells by sample in the Dimensionality Reduction Plot, and may simplify configuration of batch correction.
Options common to Loom and MEX importers
Loom and MEX formats can contain both the total expression, spliced, and unspliced counts. The importers can be configured which type of data to import and produce either an Expression Matrix (![]() ), or an Expression Matrix with spliced and unspliced counts (
), or an Expression Matrix with spliced and unspliced counts (![]() ).
).
- Import expressions. Enables import of total expression from the relevant file. This is needed when:
- spliced/unspliced counts are not available;
- the total expression of a gene cannot be obtained purely from the spliced and, if selected, unspliced counts. For example, the expression has been normalized.
- Import spliced/unspliced. Enables import of spliced and unspliced counts from the relevant file(s). If the file(s) do not contain spliced/unspliced counts, the import will fail with a relevant message.
- Include unspliced counts in total expression. By default, the total expression of a gene is obtained from the spliced counts. When this option is enabled, the unspliced counts are also added to the total expression. This option is recommended for single nucleus RNA sequencing (snRNA-Seq), where data is usually analyzed by counting expression from both exons and introns [Bakken et al., 2018]. This option has no effect when both 'Import expressions' and 'Import spliced/unspliced' are enabled, where the total expression is read directly from the file.
Details specific to the Loom importer
Loom allows the exchange of data between different software packages.
A Loom file has an internal structure consisting of a main matrix, optional `layers' of the same size as the main matrix, row and column attributes (describing features and cells, respectively), and sparse graphs describing links between features or between cells. See https://linnarssonlab.org/loompy/format/index.html for details of the format.
The Loom importer expects the Loom format version 3.0.0 and imports the main matrix, row attributes describing feature names and feature identifiers, column attributes, and, if Import spliced/unspliced is ticked, the layers containing the spliced and unspliced counts. All other information in the Loom file is ignored.
- Spliced layer. The layer where the spliced counts are stored.
- Unspliced layer. The layer where the unspliced counts are stored.
- Cell ID attribute. A column attribute identifying the cell by its barcode and sample. The interpretation of this value is specified by the Cell format.
- Gene or transcript ID attribute. A row attribute describing an identifier for a gene or transcript (e.g., ENSG00000243485 for ENSEMBL). If no identifiers are present, then it is also possible to set this to the same value as the Gene or transcript name attribute.
- Gene or transcript name attribute. A row attribute describing the name for a gene or transcript. If no names are present, then it is also possible to set this to the same value as the Gene or transcript ID attribute.
- Create clusters for. A comma-separated list of column attributes to be imported as Cell Clusters (
 ). These attributes must be string arrays. Any other column attributes will be imported as Cell Annotations (
). These attributes must be string arrays. Any other column attributes will be imported as Cell Annotations ( ).
).
- Map clusters to QIAGEN Cell Ontology. When this is enabled, clusters will be translated, if possible, to the QIAGEN Cell Ontology (see The QIAGEN Cell Ontology). The translation attempts to match each cluster with a cell type from the ontology based on the name and known synonyms. For example, `alveolar epithelial cells' are also called `pneumocytes'. If the data file contains an `alveolar epithelial cells' cluster and this option is selected, then in the imported file the cluster will be named `pneumocytes'. This option can be useful when standardizing cell annotations from different sources. It is especially recommended if the imported data will be used to extend a QIAGEN Cell Type Classifier using the Train Cell Type Classifier tool (Train Cell Type Classifier).
Details specific to the MEX importer
The MEX importer requires at least two files to be supplied:
- Barcodes file. A file with the extension .tsv and tab-separated columns, with one row per barcode. It can optionally contain a header. The barcodes are read from the first column. Empty lines are ignored.
Use the Cell format option to control how the barcodes should be interpreted - for example if it also includes information about the sample.
- Feature file. A file with the extension .tsv and one row per feature. It can optionally contain a header. Empty lines are ignored.
It contains multiple tab-separated columns:
- One column: the feature name.
- Two columns: the feature identifier and name.
- Three columns: the feature identifier, name, and type. Of the commonly used feature types, "Gene Expression", "Transcript Expression", and "Spike-in" are the most important ones. Other features, such as "Antibody Capture" will be silently ignored by most tools.
For 10x Multiome files there will be six columns. The last three consist of genome coordinates and will be ignored. Lines with feature type "Peaks" will also be ignored. They should instead be imported as a Peak Count Matrix (see Import Peak Count Matrix).
- Matrix file. A file containing the expression with the extension .mtx in the Matrix Market Exchange Coordinate Format.
- Matrix file (spliced). A file containing the spliced counts with the extension .mtx in the Matrix Market Exchange Coordinate Format.
- Matrix file (unspliced). A file containing the unspliced counts with the extension .mtx in the Matrix Market Exchange Coordinate Format.
- Name. The name of the imported matrix. If Cell format is not configured to parse a sample name from each barcode in the barcodes file, then this will also be the sample name for all the imported barcodes.
- Files are in same directory. This option is provided for convenience. When enabled, updating any one of the three files to a file in a new directory will lead to automatic updates of the other two files, if suitable candidates can be found in the same directory. This option only works for local files.
See additional details in the 'Options common to Loom and MEX importers' section on how the total expression is calculated.
See https://math.nist.gov/MatrixMarket/formats.html for details of the Matrix Market Exchange Coordinate Format.
Details specific to the MEX archive importer
The MEX archive importer is provided for convenience. It accepts a .zip, .tar or .tar.gz archive file containing the files required by the MEX importer. In order to uniquely identify each file, these must have a specific name:
- Barcodes file must be named barcodes.tsv
- Feature file must either be named features.tsv or genes.tsv
- Matrix file must be named matrix.mtx
- Matrix file (spliced) must be named spliced.mtx
- Matrix file (unspliced) must be named unspliced.mtx
The importer can be configured to either import an Expression Matrix (![]() ), or an Expression Matrix with spliced and unspliced counts (
), or an Expression Matrix with spliced and unspliced counts (![]() ). For the first option, 'Import expressions' must be enabled, while for the second option, 'Import spliced/unspliced' must be enabled.
). For the first option, 'Import expressions' must be enabled, while for the second option, 'Import spliced/unspliced' must be enabled.
Either the 'Matrix file', or 'Matrix file (spliced)' and Matrix file (unspliced)' can be missing from the archive, depending on how the importer has been configured.
See additional details in the 'Options common to Loom and MEX importers' section on how the total expression is calculated.
Details specific to the CSV importer
The CSV/TXT importer supports import of text data in a full table format.
- Table layout. Choose whether the table has cells in columns and features in rows, or is transposed such that features are in columns and cells are in rows.
- Separator. Choose the column separator.
|
Working with spreadsheets Be careful to check that all the data is present before import if the file originates from a spreadsheet program. Such programs often impose limits on the number of rows and columns. |
