Importing data
The elements needed for executing the workflows can either be imported prior to the execution, or can be automatically imported during execution, see http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Launching_workflows_individually_in_batches.
For details on importers, see Data import.
If the workflow uses two different types of input elements, such as Expression Matrix (![]() ) / (
) / (![]() ) and Peak Count Matrix (
) and Peak Count Matrix (![]() ) or Expression Matrix (
) or Expression Matrix (![]() ) / (
) / (![]() ) and TCR Cell Clonotypes (
) and TCR Cell Clonotypes (![]() ) or BCR Cell Clonotypes (
) or BCR Cell Clonotypes (![]() ), the sample in the input elements must be the same for cells originating from the same sample. This can be achieved in different ways, depending on how the elements were generated:
), the sample in the input elements must be the same for cells originating from the same sample. This can be achieved in different ways, depending on how the elements were generated:
- If the input elements were generated in the CLC Single Cell Analysis Module, the sample name can be set when running the Annotate Reads with Cell and UMI tool, see Setting the sample name.
- If the input elements are imported, the sample name can be set during import through the Cell format or Sample options, see Cell format in importers. This can be used both when importing the elements prior to or during execution.
- The tool Update to Common Sample Name (
 ) can be used for updating the sample name in either input element, see Update to Common Sample Name. This requires processing the elements before executing the workflow.
) can be used for updating the sample name in either input element, see Update to Common Sample Name. This requires processing the elements before executing the workflow.
Care must be take when executing a workflow in batch mode using on-the-fly import, as options that are explicitly supplied will be used for all imported files:
- Defining the sample through the Cell format or Sample options. If the files to be imported do not share the same Cell format and / or Sample, they need to be imported separately before the workflow execution.
- Importing nearby genes and / or transcription factors for Peak Count Matrices. The HDF5 importer is limited to reading just one file for nearby genes and/or transcription factors (figure 16.4), while the archive MEX format allows bundling separate nearby genes and/or transcription factors in each archive (figure 16.5) and is hence better suited for batch execution.
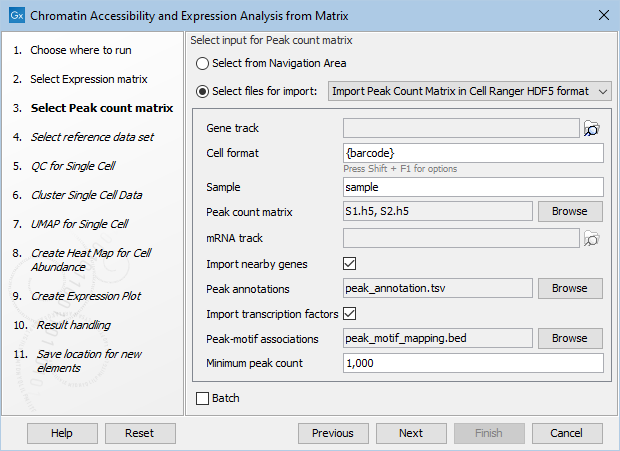
Figure 16.4: Specifying nearby genes and transcription factors for HDF5 import.
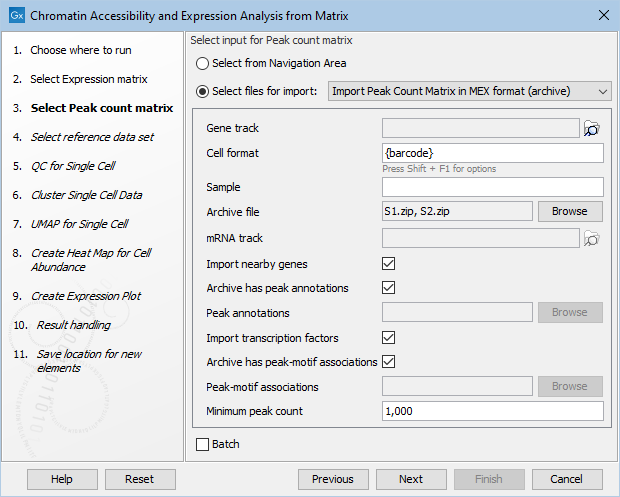
Figure 16.5: Specifying nearby genes and transcription factors for archive MEX import.
