Train Cell Type Classifier
The Train Cell Type Classifier tool trains a Cell Type Classifier which can be used in the Predict Cell Types tool (see Predict Cell Types).
The tool learns to distinguish different cell types by learning specific expression patterns from the expression values of cells that are already assigned a cell type.
It can be found in the Toolbox here:
Gene Expression (![]() ) | Cell Type Classification (
) | Cell Type Classification (![]() ) | Train Cell Type Classifier (
) | Train Cell Type Classifier (![]() )
)
The tool takes an Expression Matrix (![]() ) / (
) / (![]() ) as input. The following options can be adjusted (figure 6.3):
) as input. The following options can be adjusted (figure 6.3):
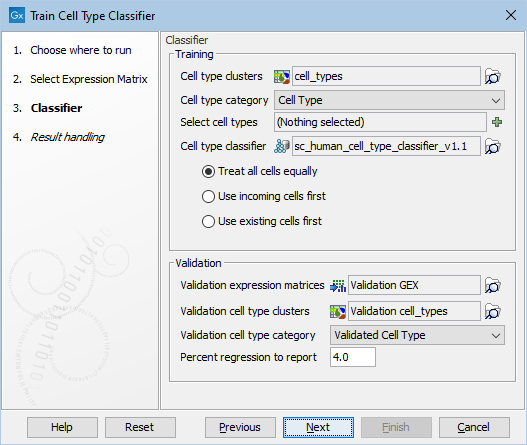
Figure 6.3: The options in the dialog of the Train Cell Type Classifier tool. A Cell Type Classifier for human data downloaded from the Reference Data Manager has been selected.
- Cell type clusters. A Cell Clusters element containing clusters for the input matrix.
- Cell type category. The category from the Cell Clusters element which contains the clusters representing cell types. The tool cannot distinguish the clusters that are true cell types, and therefore this category should only contain clusters that truly represent cell types. It is not required for all cells to belong to a cluster and cells with unknown cell types should be left unannotated, rather than being clustered in an "unknown" cluster.
- Select cell types (Optional). Only train using the selected clusters. If no cell types are selected, all will be used. This can be used to remove undesired "unknown" clusters, or to remove cell types that are found to reduce performance when added to an existing Cell Type Classifier.
- Cell type classifier (Optional). A Cell Type Classifier downloaded from the Reference Data Manager (see The Reference Data Manager) or produced by this tool. This allows extending existing classifiers with new data. To keep the running times and the size of the resulting Cell Type Classifier low, the tool uses up to approximately 50 training cells per cell type, which are chosen randomly to include cells from every sample present in the data. If the data contains more than 50 samples, one cell will be chosen randomly from each sample. When a Cell Type Classifier is provided, the cells to be used during training can be preferentially chosen from the classifier or the incoming data as follows:
- Treat all cells equally. The tool will use cells from both the classifier and the incoming data in a as uniform manner as possible. This is the default option and it ensures that all samples present in both the classifier and incoming data are represented in the training cells.
- Use incoming cells first. The tool will preferentially use cells from the incoming data. If there are less than 50 cells for any given cell type, further cells will be chosen from the classifier.
- Use existing cells first. The tool will preferentially use cells from the classifier. If there are less than 50 cells for any given cell type, further cells will be chosen from the incoming data.
- Validation expression matrices (Optional). One or more Expression Matrices (
 ) / (
) / ( ) containing cells on which to evaluate the performance of the Cell Type Classifier, and to detect regressions from the existing Cell Type Classifier if present.
The tool performs validation only for cells that are not used during training, including the data present in the existing classifier, if provided. For validation it is therefore recommended to use all existing training data that has been previously used for training a classifier.
) containing cells on which to evaluate the performance of the Cell Type Classifier, and to detect regressions from the existing Cell Type Classifier if present.
The tool performs validation only for cells that are not used during training, including the data present in the existing classifier, if provided. For validation it is therefore recommended to use all existing training data that has been previously used for training a classifier.
The report produced can be used to detect inconsistencies in the annotation between the input matrix, the data present in the existing classifier, if used, and the validation matrices.
Note that the features in the input matrix and validation matrices should be matching, see Features used for training and prediction.
- Validation cell type clusters (Optional). One or more Cell Clusters elements containing clusters for the validation matrices. It is not required for there to be a one-to-one correspondence between validation matrices and validation cell type clusters. For example, a single validation Cell Clusters element may contain clusters for some of the cells in several of the validation matrices. Only cells that are present in both a validation matrix and a validation cell type cluster are used for validation.
- Validation cell type category (Optional). The category from the Cell Clusters element(s) which contains the clusters representing cell types. The tool cannot distinguish the clusters that are true cell types, and therefore this category should only contain clusters that truly represent cell types. It is not required for all cells to belong to a cluster and cells with unknown cell types should be left unannotated, rather than being clustered in an "unknown" cluster.
- Percent regression to report (Optional). Only relevant when a Cell Type Classifier has been provided. If the threshold is
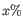 , then in the report:
, then in the report:
- Cell types with
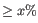 less sensitivity on the validation matrix are colored red.
less sensitivity on the validation matrix are colored red.
- Cell types with
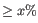 more sensitivity on the validation matrix are colored green.
more sensitivity on the validation matrix are colored green.
- Cell types with
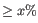 less sensitivity in any validation matrix are listed in a table.
less sensitivity in any validation matrix are listed in a table.
- For each validation matrix with
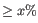 less sensitivity, a list of incorrect predictions to cell types that are present in the input matrix is produced. The list is filtered to only contain cell types that are predicted incorrectly with
less sensitivity, a list of incorrect predictions to cell types that are present in the input matrix is produced. The list is filtered to only contain cell types that are predicted incorrectly with 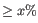 more.
more.
- Cell types with
Subsections
- Features used for training and prediction
- Interpreting the output of Train Cell Type Classifier
- SVMs for cell type classification
