Filter Cell Clonotypes
Sometimes it can be desirable to restrict TCR Cell Clonotypes (The Filter Cell Clonotypes tool can be found in the Toolbox here:
Immune Repertoire (![]() ) | Filter Cell Clonotypes (
) | Filter Cell Clonotypes (![]() )
)
The tool takes a Cell Clonotypes element as input and produces a filtered element.
The following options can be adjusted (figure 10.4):
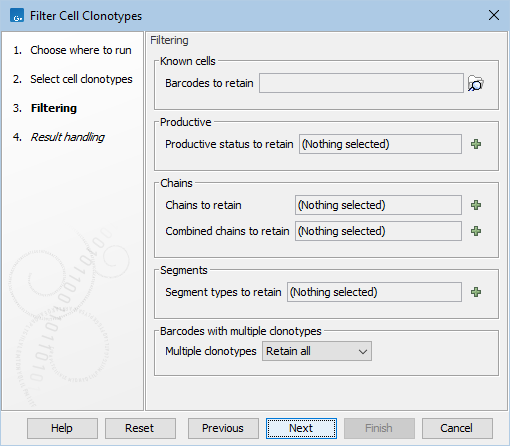
Figure 10.4: The options in the dialog of the Filter Cell Clonotypes tool.
- Barcodes to retain. Multiple elements containing cells can be provided, such as Expression Matrices, Cell Clusters and Cell Annotations. From these, a set of valid cells, identified through the sample and barcode, is obtained as the intersection of the cells in the chosen elements. When used, only the clonotypes for the valid cells are retained in the output.
- Productive status to retain. A mixture of 'Productive', 'Out of frame' and 'Premature stop codon' can be chosen and only the clonotypes with the respective productive status will be retained. If left empty, no filter is applied.
- Chains to retain. A mixture of:
- For TCR Cell Clonotypes: TRA, TRB, TRG and TRD;
- For BCR Cell Clonotypes: IGH, IGHL and IGK.
- Combined chains to retain. A mixture of:
- For TCR Cell Clonotypes: TRA + TRB and TRG + TRG;
- For BCR Cell Clonotypes: a combination of IGH, IGK and IGL with at most two heavy and two light chains. For example, IGH + IGL or IGH + IGH + IGK + IGL.
- Segment types to retain. A mixture of 'V', 'D', 'J' and 'C' can be chosen and only the clonotypes that have identified segments for all respective segment types will be retained. This means that, for example, if 'D' is chosen, only chains for which the D segment is used will be retained, and for those chains, only the clonotypes for which the identification of the D segment was successful will be retained. If left empty, no filter is applied.
- Barcodes with multiple clonotypes. Barcodes can have more than one clonotype associated with them. Ideally, for each barcode all chains have been identified that together form the complete receptor: both TRA + TRB or TRG + TRD chains for T cell, and both the two heavy (IGH) and two light (IGK or IGL) chains for B cells. Sometimes a barcode can have fewer or more clonotypes. Configuring the Multiple clonotypes option determines how such barcodes should be handled:
- Retain all. No filter is applied and all clonotypes are retained.
- Retain primary / secondary. When a barcode has more clonotypes for the same chain than needed for the complete receptor, clonotypes are marked as primary or secondary based on the number of reads. For example,
- If a T cell barcode contains two TRB chains, the chain with the highest number of reads will be part of the primary clonotype, while the chain with the lowest number of reads will be part of the secondary clonotype.
- If a B cell barcode contains three light chains, the two with the highest number of reads will be part of the primary clonotype, while the one with the lowest number of reads will be part of the secondary clonotype.
- Retain none. If a barcode contains secondary clonotypes, the entire barcode is removed. Note that Retain primary only removes the secondary clonotypes, while Retain none removes the entire barcode that contains secondary clonotypes, including its primary clonotypes.
The options above can be mixed and matched to obtain the desired output. Note that the filters are applied in the order given above.
For example, assume we want to only use the primary productive TRB clonotypes. This can be obtained by setting "Productive status to retain" to "Productive", "Chains to retain" to "TRB", and "Multiple clonotypes" to "Retain primary". If one barcode A has a primary non-productive and a secondary productive TRB clonotype, the non-productive clonotype will be removed first and the productive one will become the primary one. Hence, "Retain primary" will have no effect on this barcode. If another barcode has two productive TRB clonotypes, "Retain primary" will remove the secondary clonotype.
If the desired behavior is that barcode A should be entirely removed from the output, as its primary clonotype is not productive, the tool can be run multiple times such that the filters are applied in a different order. By running the tool with "Multiple clonotypes" to "Retain primary" first, the barcode will have only the non-productive clonotype. A second execution of the tool with "Productive status to retain" set to "Productive" and "Chains to retain" set to "TRB" will entirely remove the barcode.
The Filter Cell Clonotypes tool can optionally produce a report for each sample found in the input element, summarizing the clonotypes left after filtering. The output report includes the same information as the report produced by the Single Cell V(D)J-Seq Analysis tool, minus the assembly and trimming summaries (see The report output from Single Cell V(D)J-Seq Analysis).
To obtain a report summarizing clonotypes across samples, see Compare Single Cell Immune Repertoires.
