When a single primer region is defined
If only a single region is defined, only single primers will
be suggested by the algorithm.
After pressing the Calculate button a dialog will appear (see figure 20.7).
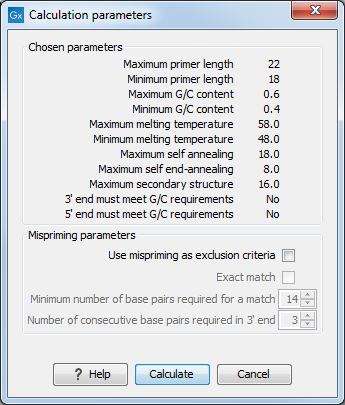
Figure 20.7: Calculation dialog for PCR primers when only a single primer region has been defined.
The top part of this dialog shows the parameter settings chosen in the Primer parameters preference group which will be used by the design algorithm.
Mispriming: The lower part contains a menu where the user can choose to include mispriming as an exclusion criteria in the design process. If this option is selected the algorithm will search for competing binding sites of the primer within the rest of the sequence, to see if the primer would match to multiple locations. If a competing site is found (according to the parameters set), the primer will be excluded.
The adjustable parameters for the search are:
- Exact match. Choose only to consider exact matches of the primer, i.e. all positions must base pair with the template for mispriming to occur.
- Minimum number of base pairs required for a match. How many nucleotides of the primer that must base pair to the sequence in order to cause mispriming.
- Number of consecutive base pairs required in 3' end. How many consecutive 3' end base pairs in the primer that MUST be present for mispriming to occur. This option is included since 3' terminal base pairs are known to be essential for priming to occur.
Note! Including a search for potential mispriming sites will prolong the search time substantially if long sequences are used as template and if the minimum number of base pairs required for a match is low. If the region to be amplified is part of a very long molecule and mispriming is a concern, consider extracting part of the sequence prior to designing primers.
