Align Protein Structure
The Align Protein Structure tool allows you to compare a protein or binding pocket in a Molecule Project with proteins from other Molecule Projects. The tool is invoked using the (![]() ) Align Protein Structure action from the Molecule Project Side Panel. This action will open an interactive dialog box (figure 13.20). By default, when the dialog box is closed with an "OK", a new Molecule Project will be opened containing all the input protein structures laid on top of one another. All molecules coming from the same input Molecule Project will have the same color in the initial visualization.
) Align Protein Structure action from the Molecule Project Side Panel. This action will open an interactive dialog box (figure 13.20). By default, when the dialog box is closed with an "OK", a new Molecule Project will be opened containing all the input protein structures laid on top of one another. All molecules coming from the same input Molecule Project will have the same color in the initial visualization.
The dialog box contains three fields:
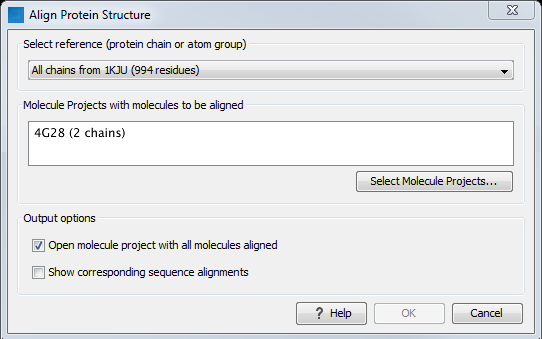
Figure 13.20: The Align Protein Structure dialog box.
- Select reference (protein chain or atom group) This drop-down menu shows all the protein chains and residue-containing atom groups in the current Molecule Project. If an atom group is selected, the structural alignment will be optimized in that area. The 'All chains from Molecule Project option will create a global alignment to all protein chains in the project, fitting e.g. a dimer to a dimer.
- Molecule Projects with molecules to be aligned One or more Molecule Projects containing protein chains may be selected.
- Output options The default output is a single Molecule Project containing all the input projects rotated onto the coordinate system of the reference. Several alignment statistics, including the RMSD, TM-score, and sequence identity, are added to the History of the output Molecule Project. Additionally, a sequence alignments of the aligned structures may be output, with the sequences linked to the 3D structure view.
Subsections
