Running part of a workflow multiple times
Where only a part of the workflow should be run multiple times, once for each input provide, or when different inputs should follow different paths in the workflow, control flow elements can be added to the workflow .
To run a part of the workflow multiple times, add an Iterate control flow element. A Collect and Distribute element can also be added downstream. All elements downstream of an Iterate element are run multiple times, until a Collect and Distribute element is encountered. The parts of the workflow downstream of the Collect and Distribute element are run only once (see Control flow elements).
For example, the workflow in figure 11.43 would run an RNA-Seq Analysis for each sample separately, and then create a single combined report for the set of samples.
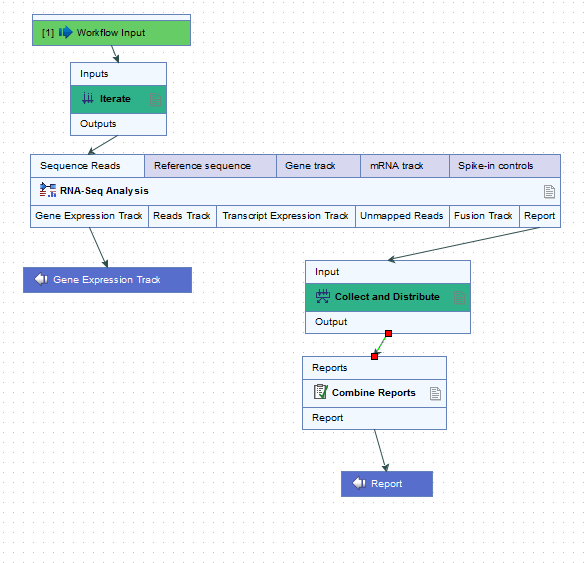
Figure 11.43: The RNA-Seq analysis tool is run once per sample and a single combined report is then generated for the full set of samples.
When running on a CLC Server the iterating parts of the workflow is run as separate jobs. For parallel execution of these iterations, job nodes or grid nodes must be available.
When using metadata table to specify the batch units, you are prompted to specify the column that defines how the samples should be grouped for execution. In figure 11.44, grouping by the column "ID" results in the RNA-Seq Analysis tool being run 8 times, once for each sample. Selecting the "Gender" column instead results in the RNA-Seq Analysis tool being run 2 times, once for each value in that column, male and female. The Combine Reports tool runs once, using information from all samples.
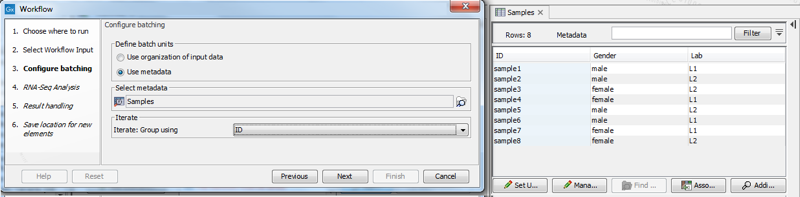
Figure 11.44: With the current selection in the wizard, the RNA-Seq Analysis tool will run 8 times, once for each sample. The Combine Reports tool will run once.
The name of workflow elements can be changed. This changes the text displayed in the wizard when the workflow is run. This can be useful if a workflow contains multiple identical elements (figure 11.45).
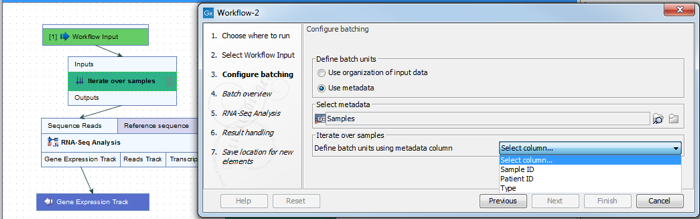
Figure 11.45: The Iterate element can be renamed to change the text that is displayed in the wizard when running the workflow.
Defining batch units when using Demultiplex Reads
When Demultiplex Reads is used in a workflow, the Group Sequences output channel is connected to an Iterate element (figure 11.46). Batch units for the iterating section of the workflow that follows, (Trim Reads, in figure 11.46), can be defined based on information provided in the barcode file imported to Demultiplex Reads, rather than a separate metadata table. For this, the CSV or Excel format file needs to contain a column with the barcodes, a column with the sample names, and further columns, containing the relevant metadata.
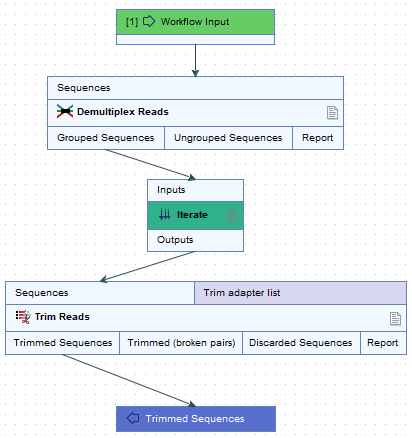
Figure 11.46: The Group Sequences output channel of Demultiplex Reads connects to an Iterate element. The data to be analyzed together in the next workflow section, i.e. the batch units, can be defined using information from the barcode file or from a separate metadata table.
