Reverse translation from protein into DNA
A protein sequence can be back-translated into DNA using CLC Genomics Workbench. Due to degeneracy of the genetic code every amino acid could translate into several different codons (only 20 amino acids but 64 different codons). Thus, the program offers a number of choices for determining which codons should be used. These choices are explained in this section. For background information see Bioinformatics explained: Reverse translation.
In order to make a reverse translation:
Toolbox | Classical Sequence Analysis (![]() ) | Protein Analysis (
) | Protein Analysis (![]() )|
Reverse Translate (
)|
Reverse Translate (![]() )
)
This opens the dialog displayed in figure 15.15:
Figure 15.15: Choosing a protein sequence for reverse translation.
If a sequence was selected before choosing the Toolbox action, the sequence is now listed in the Selected Elements window of the dialog. Use the arrows to add or remove sequences or sequence lists from the selected elements. You can translate several protein sequences at a time.
Adjust the parameters for the translation in the dialog shown in figure 15.16.
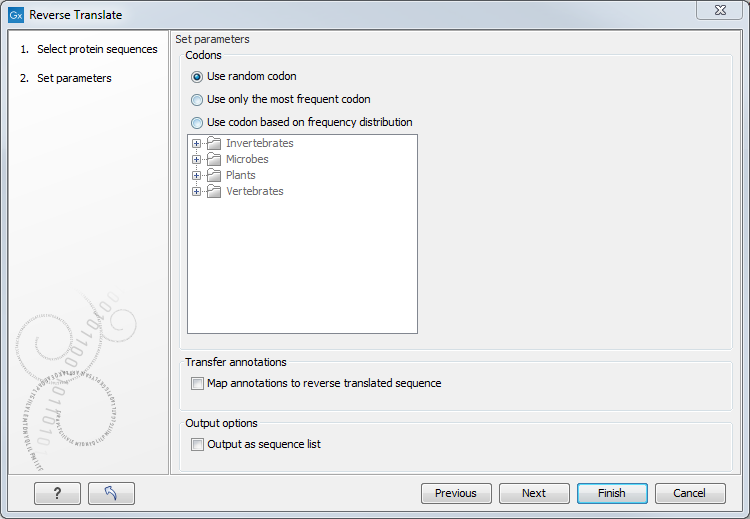
Figure 15.16: Choosing parameters for the reverse translation.
- Use random codon. This will randomly back-translate an amino acid to a codon assuming the genetic code to be 1, but without using the codon frequency tables. Every time you perform the analysis you will get a different result.
- Use only the most frequent codon. On the basis of the selected translation table, this parameter/option will assign the codon that occurs most often. When choosing this option, the results of performing several reverse translations will always be the same, contrary to the other two options.
- Use codon based on frequency distribution. This option is a mix of the other two options. The selected translation table is used to attach weights to each codon based on its frequency. The codons are assigned randomly with a probability given by the weights. A more frequent codon has a higher probability of being selected. Every time you perform the analysis, you will get a different result. This option yields a result that is closer to the translation behavior of the organism (assuming you choose an appropriate codon frequency table).
- Map annotations to reverse translated sequence. If this checkbox is checked, then all annotations on the protein sequence will be mapped to the resulting DNA sequence. In the tooltip on the transferred annotations, there is a note saying that the annotation derives from the original sequence.
The Codon Frequency Table is used to determine the frequencies of the codons. Select a frequency table from the list that fits the organism you are working with. A translation table of an organism is created on the basis of counting all the codons in the coding sequences. Every codon in a Codon Frequency Table has its own count, frequency (per thousand) and fraction which are calculated in accordance with the occurrences of the codon in the organism. The tables provided were made using Codon Usage database http://www.kazusa.or.jp/codon/ that was built on The NCBI-GenBank Flat File Release 160.0 [June 15 2007]. You can customize the list of codon frequency tables for your installation, see Custom codon frequency tables.
Click Finish to start the tool.
The newly created nucleotide sequence is shown, and if the analysis was performed on several protein sequences, there will be a corresponding number of views of nucleotide sequences.
Subsections
