Output from primer design
The output generated by the primer design algorithm is a table of proposed primers or primer pairs with the accompanying information (see figure 16.6).
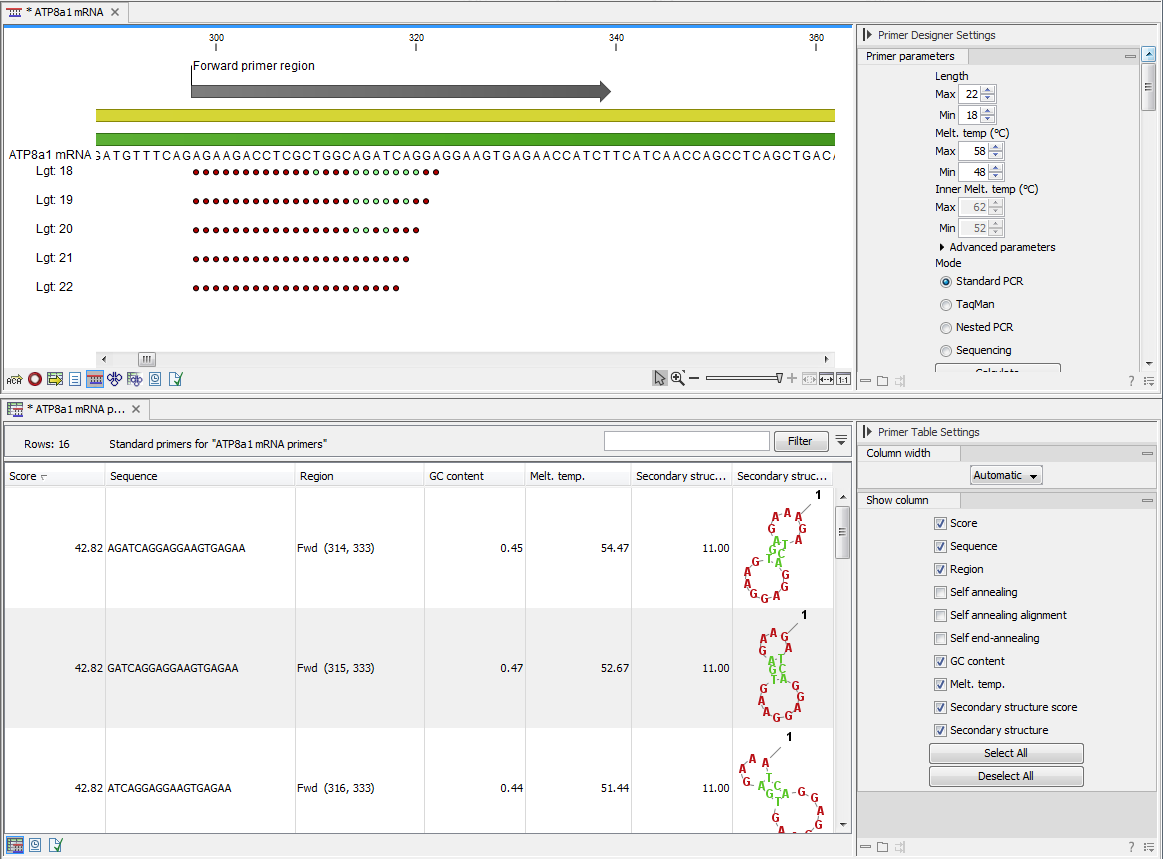
Figure 16.6: Proposed primers.
In the preference panel of the table, it is possible to customize which columns are shown in the table. See Standard PCR, Nested PCR, TaqMan or Sequencing primers for a description of the available information.
The columns in the output table can be sorted by the present information. For example the user can choose to sort the available primers by their score (default) or by their self annealing score, simply by right-clicking the column header.
The output table interacts with the accompanying primer editor such that when a proposed combination of primers and probes is selected in the table the primers and probes in this solution are highlighted on the sequence.
Saving primers Primer solutions in a table row can be saved by selecting the row and using the right-click mouse menu. This opens a dialog that allows the user to save the primers to the desired location. Primers and probes are saved as DNA sequences in the program. This means that all available DNA analyzes can be performed on the saved primers. Furthermore, the primers can be edited using the standard sequence view to introduce e.g. mutations and restriction sites.
Saving PCR fragments The PCR fragment generated from the primer pair in a given table row can also be saved by selecting the row and using the right-click mouse menu. This opens a dialog that allows the user to save the fragment to the desired location. The fragment is saved as a DNA sequence and the position of the primers is added as annotation on the sequence. The fragment can then be used for further analysis and included in e.g. an in-silico cloning experiment using the cloning editor.
Adding primer binding annotation You can add an annotation to the template sequence specifying the binding site of the primer: Right-click the primer in the table and select Mark primer annotation on sequence.
