Add attB sites
The first step in the Gateway cloning process is to amplify the target sequence with primers including so-called attB sites:
Toolbox | Molecular Biology Tools (![]() ) | Cloning and Restriction Sites (
) | Cloning and Restriction Sites (![]() )| Gateway Cloning (
)| Gateway Cloning (![]() ) | Add attB Sites (
) | Add attB Sites (![]() )
)
This will open a dialog where you can select one or more sequences. Note that if your fragment is part of a longer sequence, you will need to extract it prior to starting the tool: select the relevant region (or an annotation) of the original sequence, right-click the selection and choose to Open Annotation in New View. Save (![]() ) the new sequence in the Navigation Area.
) the new sequence in the Navigation Area.
When you have selected your fragment(s), click Next.
This will allow you to choose which attB sites you wish to add to each end of the fragment as shown in figure 18.29.
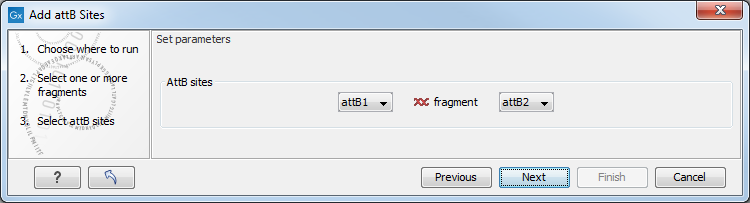
Figure 18.29: Selecting which attB sites to add.
The default option is to use the attB1 and attB2 sites. If you have selected several fragments and wish to add different combinations of sites, you will have to run this tool once for each combination.
Next, you are given the option to extend the fragment with additional sequences by extending the primers 5' of the template-specific part of the primer, i.e., between the template specific part and the attB sites.
You can manually type or paste in a sequence of your choice, but it is also possible to click in the text field and press Shift + F1 (Shift + Fn + F1 on Mac) to show some of the most common additions (see figure 18.30). Use the up and down arrow keys to select a tag and press Enter. To learn how to modify the default list of primer additions, see Extending the pre-defined list of primer additions.
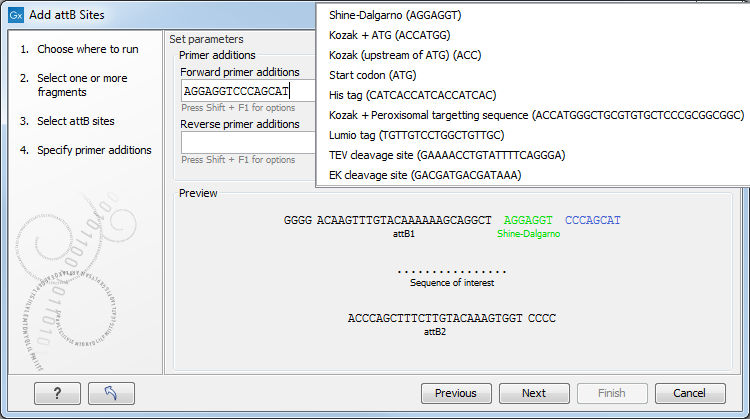
Figure 18.30: Primer additions 5' of the template-specific part of the primer where a Shine-Dalgarno site has been added between the attB site and the gene of interest.
At the bottom of the dialog, you can see a preview of what the final PCR product will look like. In the middle there is the sequence of interest. In the beginning is the attB1 site, and at the end is the attB2 site. The primer additions that you have inserted are shown in colors.
In the next step, specify the length of the template-specific part of the primers as shown in figure 18.31.
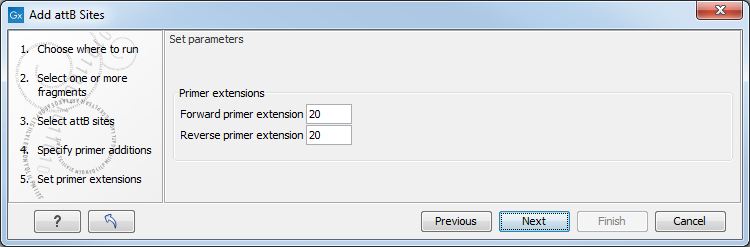
Figure 18.31: Specifying the length of the template-specific part of the primers.
CLC Genomics Workbench is not doing any kind of primer design when adding the attB sites. As a user, you simply specify the length of the template-specific part of the primer, and together with the attB sites and optional primer additions, this will be the primer. The primer region will be annotated in the resulting attB-flanked sequence. You can also choose to get a list of primers in the Result handling dialog (see figure 18.32).
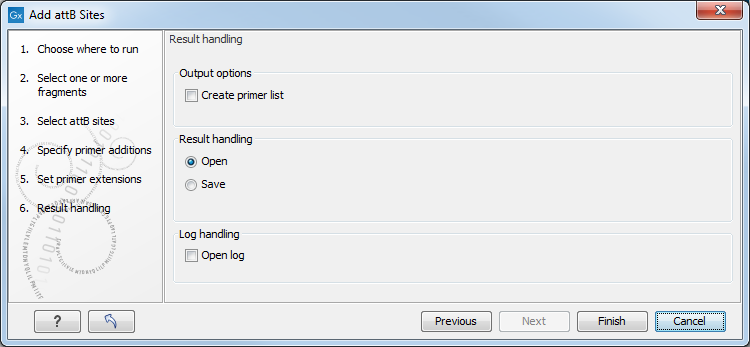
Figure 18.32: Besides the main output which is a copy of the input sequence(s) now including attB sites and primer additions, you can get a list of primers as output.
The attB sites, the primer additions and the primer regions are annotated in the final result as shown in figure 18.33 (you may need to switch on the relevant annotation types to show the sites and primer additions).

Figure 18.33: the attB site plus the Shine-Dalgarno primer addition is annotated.
There will be one output sequence for each sequence you have selected for adding attB sites. Save (![]() ) the resulting sequence as it will be the input to the next part of the Gateway cloning workflow (see Create entry clones(BP)).
) the resulting sequence as it will be the input to the next part of the Gateway cloning workflow (see Create entry clones(BP)).
Subsections
