Bioinformatics explained: Proteolytic cleavage
Proteolytic cleavage is basically the process of breaking the peptide bonds between amino acids in proteins. This process is carried out by enzymes called peptidases, proteases or proteolytic cleavage enzymes.
Proteins often undergo proteolytic processing by specific proteolytic enzymes (proteases/peptidases) before final maturation of the protein. Proteins can also be cleaved as a result of intracellular processing of, for example, misfolded proteins. Another example of proteolytic processing of proteins is secretory proteins or proteins targeted to organelles, which have their signal peptide removed by specific signal peptidases before release to the extracellular environment or specific organelle.
Below a few processes are listed where proteolytic enzymes act on a protein substrate.
- N-terminal methionine residues are often removed after translation.
- Signal peptides or targeting sequences are removed during translocation through a membrane.
- Viral proteins that were translated from a monocistronic mRNA are cleaved.
- Proteins or peptides can be cleaved and used as nutrients.
- Precursor proteins are often processed to yield the mature protein.
Proteolytic cleavage of proteins has shown its importance in laboratory experiments where it is often useful to work with specific peptide fragments instead of entire proteins.
Proteases also have commercial applications. As an example proteases can be used as detergents for cleavage of proteinaceous stains in clothing.
The general nomenclature of cleavage site positions of the substrate were formulated by Schechter and Berger, 1967-68 [Schechter and Berger, 1967], [Schechter and Berger, 1968]. They designate the cleavage site between P1-P1', incrementing the numbering in the N-terminal direction of the cleaved peptide bond (P2, P3, P4, etc..). On the carboxyl side of the cleavage site the numbering is incremented in the same way (P1', P2', P3' etc. ). This is visualized in figure 15.21.
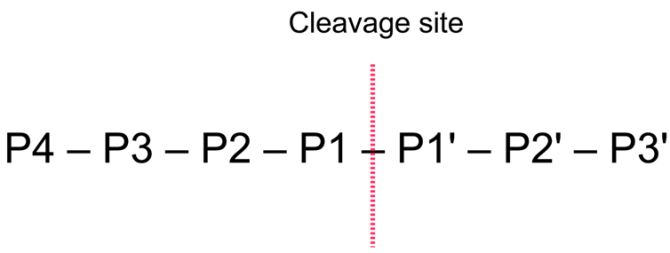
Figure 15.21: Nomenclature of the peptide substrate. The substrate is cleaved between position P1-P1'.
Proteases often have a specific recognition site where the peptide bond is cleaved. As an example trypsin only cleaves at lysine or arginine residues, but it does not matter (with a few exceptions) which amino acid is located at position P1'(carboxyterminal of the cleavage site). Another example is trombin which cleaves if an arginine is found in position P1, but not if a D or E is found in position P1' at the same time. (See figure 15.22).
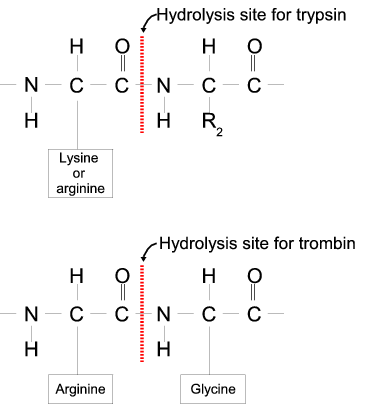
Figure 15.22: Hydrolysis of the peptide bond between two amino acids.
Trypsin cleaves unspecifically at lysine or arginine residues whereas trombin cleaves at arginines if asparate or glutamate is absent.
Bioinformatics approaches are used to identify potential peptidase cleavage sites. Fragments can be found by scanning the amino acid sequence for patterns which match the corresponding cleavage site for the protease. When identifying cleaved fragments it is relatively important to know the calculated molecular weight and the isoelectric point.
Other useful resources
The Peptidase Database: http://merops.sanger.ac.uk/
