Output from the Detect QIAseq RNAscan Fusions workflow
The following outputs are generated:
- Gene expression (
 ): A track with gene expression annotations, including counts and expression values for each gene. See RNA-Seq result handling for details.
): A track with gene expression annotations, including counts and expression values for each gene. See RNA-Seq result handling for details.
- Genome Browser View (
 ): A collection of output tracks, allowing multiple data types at the same genomic position to be viewed simultaneously. Note that not all output tracks are necessarily in the browser. Tracks can be added and removed from the browser. Two browsers are generated, one for wild type (WT) and one for the fusion chromosomes.
): A collection of output tracks, allowing multiple data types at the same genomic position to be viewed simultaneously. Note that not all output tracks are necessarily in the browser. Tracks can be added and removed from the browser. Two browsers are generated, one for wild type (WT) and one for the fusion chromosomes.
- PASS fusion genes (WT) (
 ): The breakpoints on the reference genome of detected fusions that have passed all relevant filters. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
): The breakpoints on the reference genome of detected fusions that have passed all relevant filters. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
- Sample report (
 ): A report containing essential information from all reports produced by the workflow. See Create Sample Report output for details.
): A report containing essential information from all reports produced by the workflow. See Create Sample Report output for details.
- QC & Reports folder:
- Create UMI reads report (
 ): Summarizes the identified UMI groups. See Create UMI Reads from Reads for details.
): Summarizes the identified UMI groups. See Create UMI Reads from Reads for details.
- Fusion report (
 ): Summarizes the identified fusions. See Output from Detect and Refine Fusion Genes for details.
): Summarizes the identified fusions. See Output from Detect and Refine Fusion Genes for details.
- QC for RNAscan Panels report (
 ): Summarizes various statistics of the mapped reads. Two reports are generated, one for the mapping of all reads ('WT') and one for the mapping of 'Fusion' reads only. See The QIAseq RNAscan Panels Report for details.
): Summarizes various statistics of the mapped reads. Two reports are generated, one for the mapping of all reads ('WT') and one for the mapping of 'Fusion' reads only. See The QIAseq RNAscan Panels Report for details.
- QC report (
 ): Summarizes and visualizes various statistics of the input reads. See QC for Sequencing Reads for details.
): Summarizes and visualizes various statistics of the input reads. See QC for Sequencing Reads for details.
- Remove and annotate UMI report (
 ): Summarizes the identified UMIs. See Remove and Annotate with Unique Molecular Index for details.
): Summarizes the identified UMIs. See Remove and Annotate with Unique Molecular Index for details.
- Remove ligation artifacts report (
 ): Summarizes ligation artifacts found in and removed from the read mapping. See Remove Ligation Artifacts for details.
): Summarizes ligation artifacts found in and removed from the read mapping. See Remove Ligation Artifacts for details.
- RNA-Seq report (
 ): Summarizes various mapping statistics and biotypes distributions. See RNA-Seq report for details.
): Summarizes various mapping statistics and biotypes distributions. See RNA-Seq report for details.
- Trim adapters, Trim homopolymers, and Trim on quality reports (
 ): Summarize the performed trimming. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Trim_output.html for details. The order of these three trimming steps can be seen in the Sample report.
): Summarize the performed trimming. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Trim_output.html for details. The order of these three trimming steps can be seen in the Sample report.
- Create UMI reads report (
- Tracks folder:
- WT subfolder containing:
- Fusion genes (WT) (
 ): The breakpoints on the reference genome of all detected fusions. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
): The breakpoints on the reference genome of all detected fusions. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
- Read mapping (
 ): The reads mapped to the reference genome. See RNA-Seq result handling for details.
): The reads mapped to the reference genome. See RNA-Seq result handling for details.
- Read mapping refined (WT) (
 ): The reads that mapped best to the reference genome. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
): The reads that mapped best to the reference genome. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
- Unaligned ends (WT) (
 ): The unaligned ends mapped to the reference genome. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
): The unaligned ends mapped to the reference genome. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
- Fusion genes (WT) (
- Fusions subfolder containing:
- Fusion genes (fusions) (
 ): The breakpoints on the artificial fusion chromosomes of all detected fusions. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
): The breakpoints on the artificial fusion chromosomes of all detected fusions. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
- Panel primers (fusions) (
 ): Information about each primer and its read coverage. See QC for RNAscan Panels for details.
): Information about each primer and its read coverage. See QC for RNAscan Panels for details.
- Read mapping (fusions) (
 ): The reads that mapped best to the artificial fusion chromosomes. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
): The reads that mapped best to the artificial fusion chromosomes. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
- Reference sequence (fusions) (
 ) , Genes (fusions) (
) , Genes (fusions) ( ), mRNA (fusions) (
), mRNA (fusions) ( ), CDS (fusions) (
), CDS (fusions) ( ), and Primers (fusions) (
), and Primers (fusions) ( ): The reference sequence, gene regions, mRNA transcripts, CDS regions, and panel primer regions corresponding to the detected fusions on the artificial fusion chromosomes. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
): The reference sequence, gene regions, mRNA transcripts, CDS regions, and panel primer regions corresponding to the detected fusions on the artificial fusion chromosomes. See https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html for details.
- Fusion genes (fusions) (
- WT subfolder containing:
Reviewing the results
The easiest way to review the results is to open the Genome Browser Views, as shown in figure 13.1.
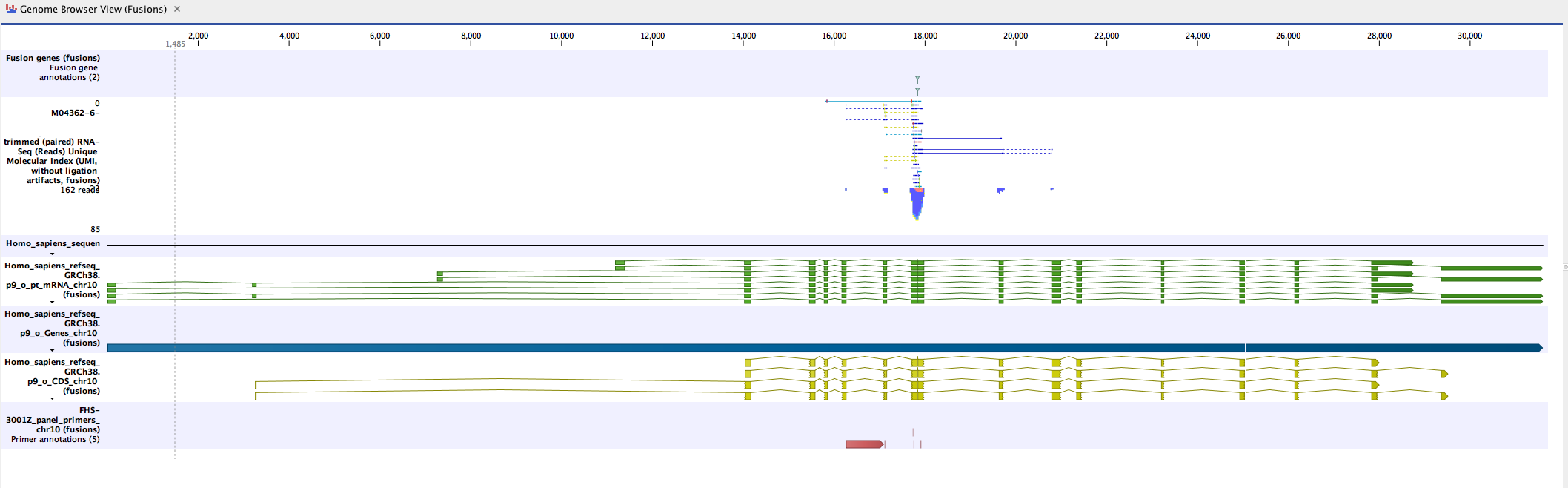
Figure 13.1: An example of a fusion in a Genome Browser View.
Double-click on the fusion track name (to the left of the Genome Browser View). The fusion track will open as a table in split view, below the Genome Browser View. Clicking on a fusion event in the table will zoom in to its location in the read mapping, allowing you to review the reads supporting the detected fusion.
Each line in the table corresponds to a fusion breakpoint, such that a fusion event is represented by two lines in the table. The two lines are linked by sharing the same 'Fusion number', which identifies the fused genes, and the same 'Fusion pair', which identifies the event for the gene. For more details on the table, see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Output_from_Detect_Refine_Fusion_Genes.html.
We recommend evaluating the support for identified fusions, see Interpretation of fusion results for details.
