Alignments for Clonotypes
The alignments view (
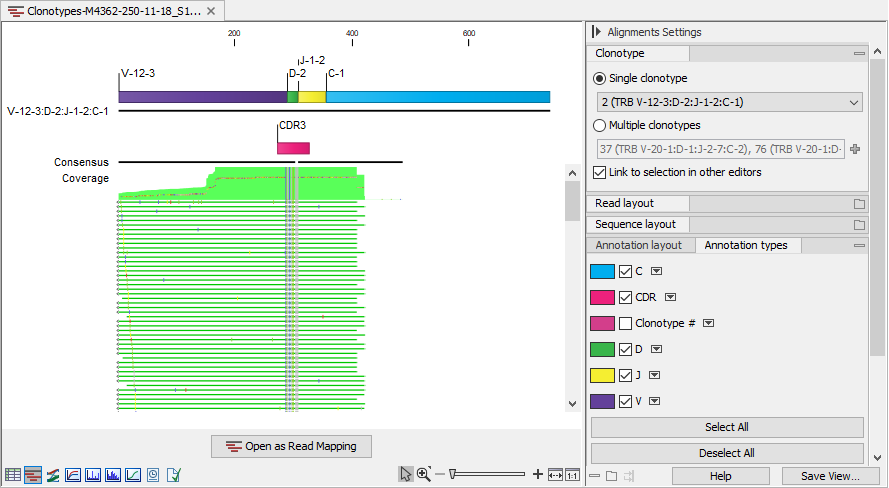
Figure 7.14: Read mapping for a TRB clonotype. V, D, J and C segments are annotated on the reference sequence and the CDR3 is annotated on the consensus.
The alignment contains:
- The reference sequence consisting of the identified V(D)JC segments. Annotations indicate the location of the different segment types. For clonotypes with ambiguous segments, the first segment from the list is used.
- The consensus sequence with an annotation indicating the CDR3 region.
- The aligned reads.
Various settings can be configured in the side panel, see https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=View_settings_in_Side_Panel.html.
For further processing, the alignments can be opened and saved as a stand-alone read mapping by using the "Open as Read Mapping" button.
The clonotypes for which the alignment should be shown can be selected from the drop-down menus in the side panel, or from the clonotype table (Clonotypes Table) while using a split view, see figure 7.15.
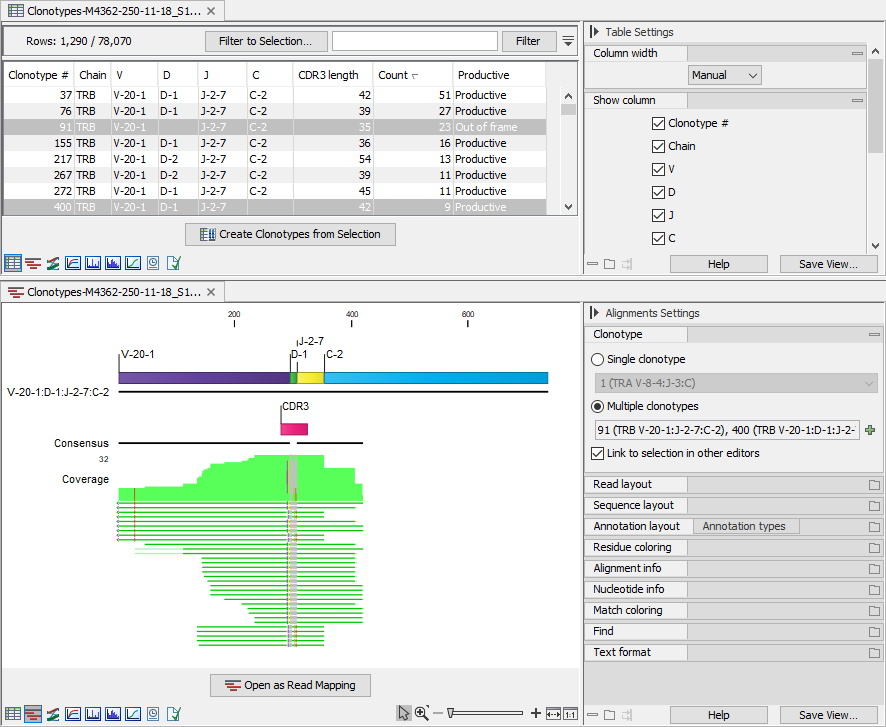
Figure 7.15: Clonotypes split view. Top: Multiple clonotypes sharing the reference segments are selected in the table view. Bottom: Alignment view for the clonotypes selected in the table view.
Alignments for multiple clonotypes can be shown together provided that they have the same V and J segments and the D / C segments are not contradictory: either the D / C segment is identified and the same, or it is missing (figure 7.15).
When viewing alignments for multiple clonotypes, it can be useful to change "Compactness" to "Not compact" and tick the "Clonotype #" annotation from the side panel. This way, it is easy to see the clonotype assigned to each read, figure 7.16.
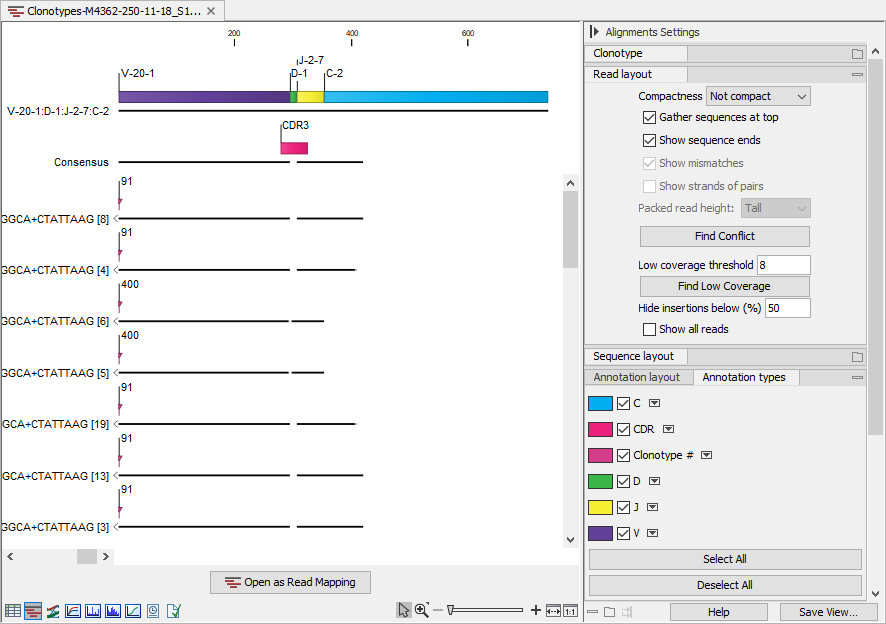
Figure 7.16: Alignment view for multiple clonotypes where "Compactness" is set to "Not compact" and "Clonotype #" is ticked.
Figure 7.15 shows an alignment for two clonotypes. Some of the reads do not span past the J segment and using the "Clonotype #" annotation, we can confirm that these reads belong to clonotype # 400, which is the clonotype without the C segment (figure 7.16). The two clonotypes do not share the D segment and have different CDR3 sequences. Using the alignment view (figure 7.15), it is straightforward to spot the differences between the two CDR3 sequences.
Alignments after merging
Alignments for clonotypes with ambiguous segments are calculated using the first segment in the list. Thus when ambiguous segments have been resolved using Merge Immune Repertoire, some reads may not be included in the alignments view. This happens when the unique segment that the ambiguous segments resolved to was not the first segment in the list.
Two situations can arise as illustrated in figure 7.17:
- The entire alignment cannot be determined. In this case, the alignment view shows an alignment of coverage 1, with one read that is identical to the reference (top right alignment in figure 7.17). This can happen when the most frequent clonotype had ambiguous segments, and ambiguity has been resolved using a less frequent clonotype. This alignment should not be used.
- The alignment for some reads cannot be determined. In this case, the alignment view does not include the reads for which the alignment could not be determined. This is apparent when the alignment coverage is lower than the clonotype count (bottom alignment in figure 7.17). This can happen when a less frequent clonotype had ambiguous segments, and ambiguity has been resolved using the most frequent clonotype. If the percentage of discarded reads is low, it will not impact the consensus sequence of the alignment.
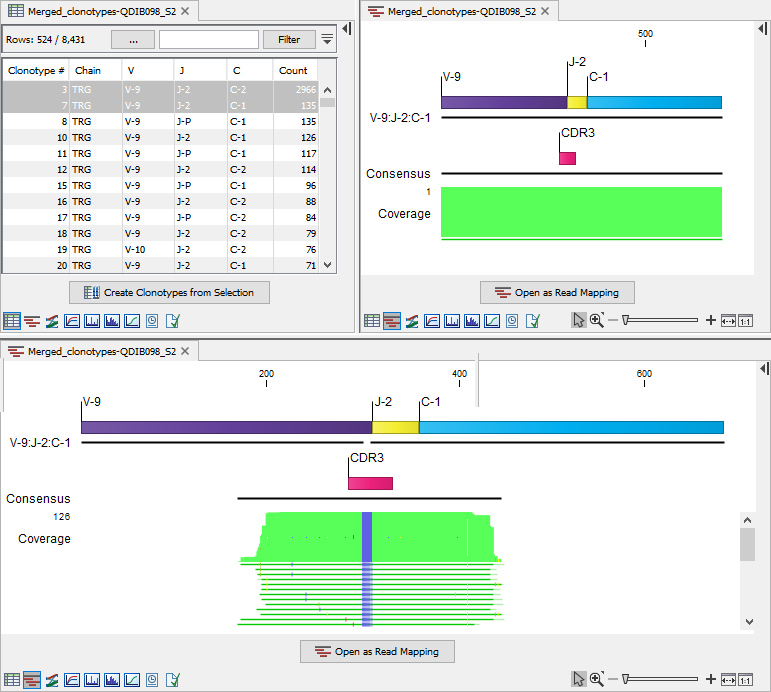
Figure 7.17: Clonotypes split view. Top left: Table view of TRG clonotypes that have been merged. Top right: Alignment view for clonotype 3. The alignment could not be determined and the view shows an alignment of coverage 1. Bottom: Alignment view for clonotype 7. The clonotype count is 135, while the alignment coverage is only 126, i.e. the alignment for 9 reads could not be determined.
