Annotate Fusions with Known Fusion Information
The Annotate Fusions with Known Fusion Information tool annotates a fusion track (on wild type chromosomes only) with information from a fusion information file given as a feature track.
Annotate Fusions with Known Fusion Information is available from the Tools menu at:
Tools | Biomedical Genomics Analysis (![]() ) | QIAseq Tools (
) | QIAseq Tools (![]() ) | Annotate Fusions with Known Fusion Information (
) | Annotate Fusions with Known Fusion Information (![]() )
)
Specify a fusion track as input (figure 5.7).
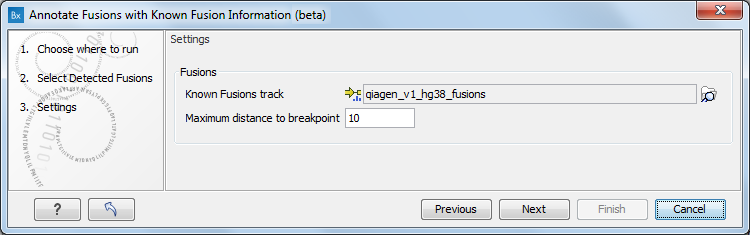
Figure 5.7: Select a fusion track, and set up the maximum distance to breakpoint.
In the next dialog, select the known fusion track you would like to use, for example the one that is saved in the CLC_References folder of the Navigation Area when downloading the QIAseq RNAscan Panels hg38 Reference Data Set for annotating with the wild type genome. You can also use a customized track imported using Import Known Fusion Information Track.
In this dialog you can also set up the maximum distance to breakpoint: the tool will annotate only fusion breakpoints for which the distance between the detected breakpoint and the closest known fusion is smaller than the one specified.
The tool outputs a fusion track. It is similar to the one that was input, but includes a new Known Fusion column, indicating the Fusion ID Number of the matching fusion in the known fusion track, or -1. Additional information from the known fusion track if also added. For example, when using the known fusion track included in the QIAseq RNAscan Panels hg38 Reference Data Set, the output track will have two more columns for Catalog Panels IDs and Cancer Types.
