Quantify miRNA
The Quantify miRNA tool counts and annotates miRNAs using miRBase and performs expression analysis of the results. The tool will output two expression tables. The "Grouped on mature" table has a row for each mature miRNA. The same mature miRNA may be produced from different precursor miRNAs. The "Grouped on seed" table has a row for each seed sequence. The same seed sequence may be found in different mature miRNAs.
The tool will take:
- Trimmed reads (both UMI or normal)
- a miRBase database
- and optionally Spike-ins: A list of sequences that have been spiked-in. Mapping against this set of sequences will be preformed before mapping of the reads against miRBase and other databases. The spike-ins are counted as exact matches and stored in the report for further analysis by the Combined miRNA Report tool.
To run the tool, go to:
Tools | RNA-Seq Analysis | miRNA Analysis (![]() ) | Quantify miRNA (
) | Quantify miRNA (![]() )
)
First select the trimmed reads as in figure 9.15.
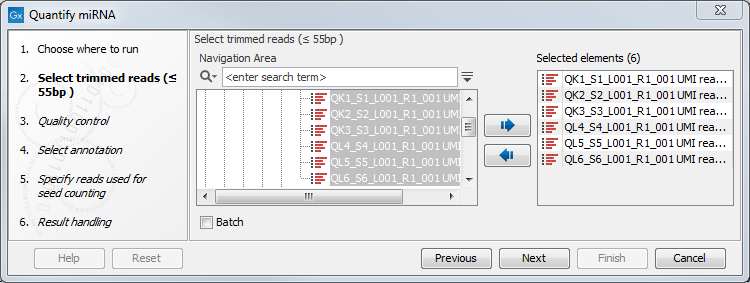
Figure 9.15: Select the reads.
If the sequencing was performed using Spike-ins controls, the option "Enable spike-ins" can be enabled in the Quality control dialog (figure 9.16), and a spike-ins file can be specified. You can also change the Low expression "Minimum supporting count", i.e., the minimum number of supporting reads for a small RNA to be considered expressed.
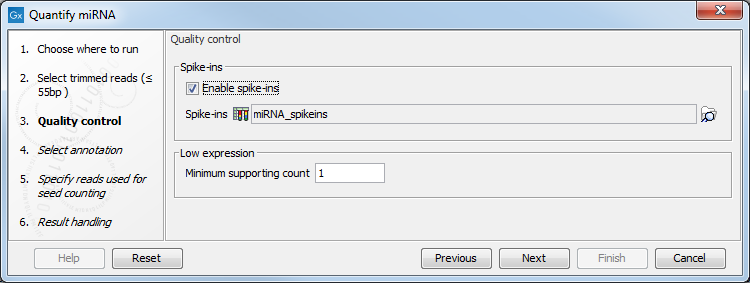
Figure 9.16: Specifying spike-ins is optional, and you can change the threshold under which a small RNA will be considered expressed.
In the annotation dialog, several configurations are available.
In the miRBase annotations section, specify a single reference - miRBase in most cases, which can be downloaded using the Download miRBase tool described here:http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Downloading_miRBase.html.
Once miRBase has been selected, click the green plus sign (note that it can take a while before all species is loaded) to see the list of species available. Species to be used for annotation should be specified using the left and right arrows, and prioritized using the up and down arrows (figure 9.17), with the species sequenced always selected as the top prioritized. The naming of the miRNA will depend on this prioritization.
In addition, it is possible to configure how specific the association between the isomiRs and the reads has to be by allowing mismatches and additional or missing bases upstream and downstream of the isomiR.
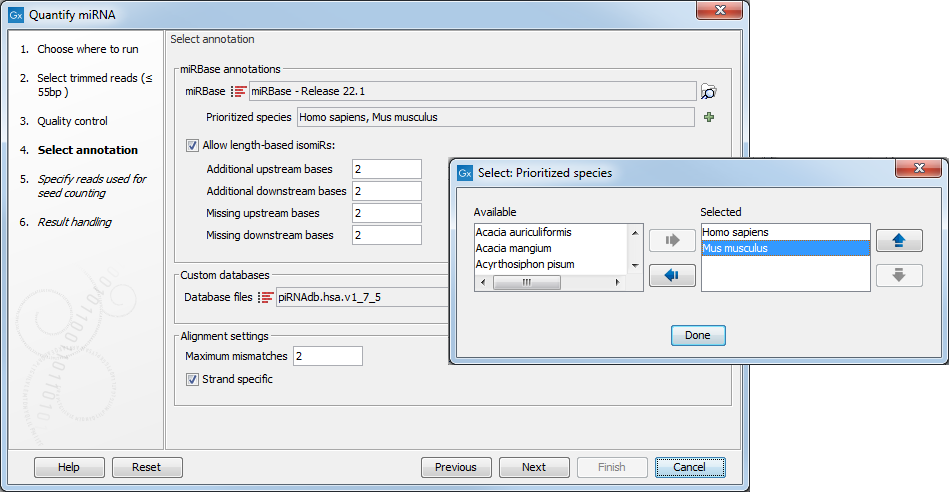
Figure 9.17: Specify and prioritize species to use for annotation, and how stringent the annotation should be.
In the Custom databases, you can optionally add sequence lists with additional smallRNA reference databases e.g. piRNAs, tRNAs, rRNAs, mRNAs, lncRNAs. An output with quantification against the custom databases can be generated, which can be used for subsequent expression analyses.
Finally, configure the Alignment settings by defining how many "Maximum mismatches" are allowed between the reads and the references, i.e. miRBase and custom databases. The option "Strand specific" is checked by default, which means that only the plus strand of the reference will be searched.
In the next dialog (figure 9.18), specify the length of the reads used for seed counting. Reads of the specified length, corresponding to the length of mature miRNA (18-25 bp by default, but this parameter can be configured) are used for seed counting. The seed is a 7 nucleotide sequence from positions 2-8 on the mature miRNA. The "Grouped on seed" output table includes a row for every seed that is observed in miRBase together with the expression of the same seed in the sample. In addition, the 20 most highly expressed novel seeds are output in the report.
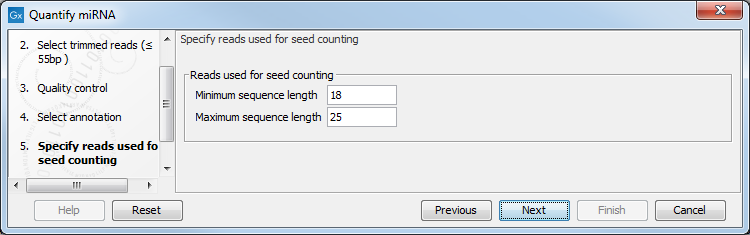
Figure 9.18: This dialog defines the length of the reads that will be merged according to their seed.
The tool will output expression tables: One is called "grouped on mature", with the miR of the mature product used as key. The other is "grouped on seed" and the sequence is used as key. See QIAseq miRNA Quantification outputs for a detailed description of the Quantify miRNA outputs. In addition, and depending on the options selected in the last dialog, the tool can output a report, a sequence list of reads that could not be mapped to any references and an expression table "grouped on custom databases" (if any was provided) that can be used for subsequent expression analysis tools such as Differential Expression, PCA for RNA-Seq and Create Heat-Map for RNA-Seq.
Naming isomiRs
The names of aligned sequences in mature groups adhere to a naming convention that generates unique names for all isomiRs. This convention is inspired by the discussion available here: https://github.com/miRTop/incubator/blob/master/isomirs/isomir_naming.md
Deletions are in lowercase and there is a suffix s for 5' (figure 9.19):

Figure 9.19: Naming of deletions.
Insertions are in uppercase and there is a suffix s for 5' (figure 9.20):

Figure 9.20: Naming of insertions.
Note that indels within miRNAs are not supported.
Mutations (SNVs) are indicated with reference symbol, position and new symbol. Consecutive mutations will not be merged into MNVs. The position is relative to the reference, so preceding (5') indels will not offset it (figure 9.21):

Figure 9.21: Naming of mutations.
Deletions followed by insertions will be annotated as in figure 9.22:

Figure 9.22: Naming of deletions followed by insertions.
If a read maps to multiple miRBase sequences we will add the suffix `ambiguous' to its name. This can happen when multiple species are selected, as they will often share the same miRBase sequence, or when a read does not map perfectly to any miRBase sequence, but it is close to two or more, distinguished by just one SNV for example.
