Start a QIAseq sample analysis
The Analyze QIAseq Samples guide offers an easy way to launch template workflows optimized for the type of analysis being run. There are several tabs, one for each application category. Under each tab, all relevant QIAseq analysis workflows are listed. For Targeted DNA, TMB/MSI Panels and RNAscan click on the arrow to the right of a workflow name to select the specific analysis type and reveal the Run button (figure 5.8).
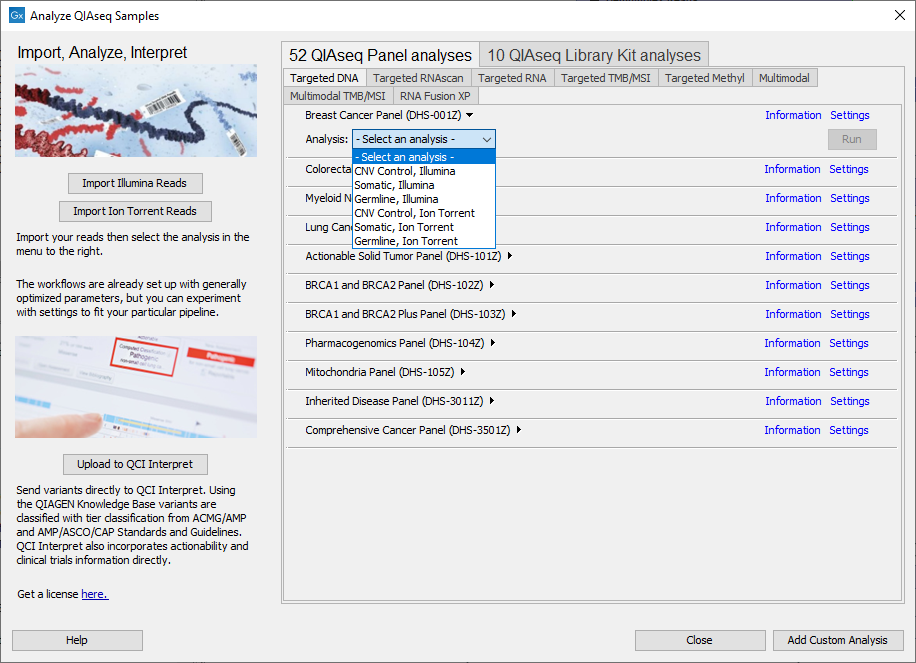
Figure 5.8: Selecting the relevant sample analysis.
Once you are ready to start a particular workflow, click on the Run button.
If you are connected to a CLC Server via your Workbench, you will be asked where you would like to run the analysis. We recommend that you run the analysis on a CLC Server when possible. Click Next.
You can then select the sequencing reads that should be analyzed (figure 5.9). The workflow can be run in batch mode by click in the Batch box. If multiple sequence lists are selected and the workflow is launched in batch mode, each sequence list would be treated as a batch unit, that is, each list would be used individually as the input for an independent run of the workflow. If multiple sequence lists are selected and the Batch box is not checked, all the lists would be used together as input for a single run of the workflow.
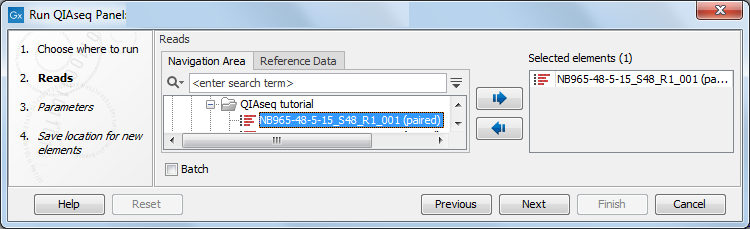
Figure 5.9: Select the sequencing reads.
If you have not done so before, you will be prompted to download the appropriate reference data (figure 5.10).
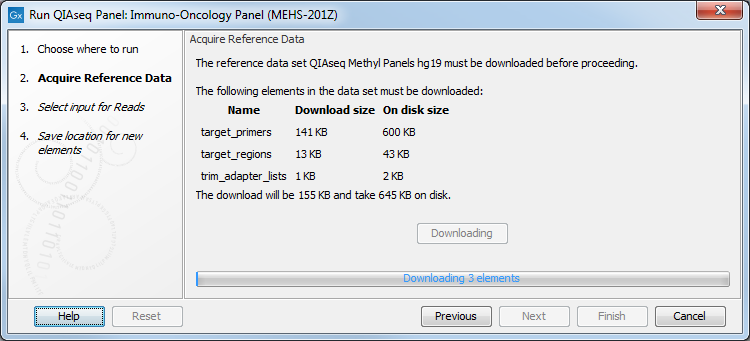
Figure 5.10: Parameters to configure for Targeted DNA workflows, here set at the default values for somatic variant detection.
The Reference Data Sets relevant to particular applications are:
- QIAseq DNA Panels hg19 for Targeted DNA workflows
- QIAseq RNAscan Panels hg38 for Targeted RNAscan workflows
- QIAseq RNA Panels hg38 for Targeted RNA workflows (hg38 or Mouse depending on the panel you want to use)
- QIAseq TMB Panels hg38 for Targeted TMB/MSI applications
- QIAseq Methyl Panels hg19 or hg38 for Targeted Methyl workflows
- QIAseq UPX Panels hg38 for UPX 3' RNA applications
- QIAseq Multimodal Panels hg38 for Multimodal applications
- QIAseq Multimodal Pan Cancer hg38 for Multimodal TMB/MSI applications
- QIAseq Exome Panels hg38 for Exome workflows
- QIAseq Immune Repertoire Analysis
- QIAseq RNA Fusion XP Panels hg38
Depending on the application, there may be parameters that can be configured. For example, for Targeted DNA panels the parameters are as shown in figure 5.11.
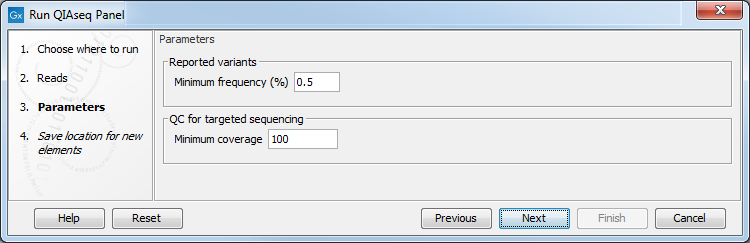
Figure 5.11: Parameters to configure for Targeted DNA workflows, here set at the default values for somatic variant detection.
- Reported variants minimum frequency The frequency threshold a variant must exceed to be included in the filtered variant track output by the workflow. For Targeted DNA panel analyses, the default value for somatic variant detection is 0.5% and for germline variant detection, it is 20%. These values can be adjusted. The lowest value with any potential effect is the minimum frequency value used when producing the original, unfiltered variant track.
All variants found will be output in the Unfiltered variants track, while only the variants passing a series of filtering, including this configurable parameter, will be output in the Variant passing filters track.
- QC for targeted sequencing minimum coverage This value will be used in the Coverage Report output by the workflow: it is set at 100 for somatic workflows, at 30 for germlines workflows
In the last dialog, choose where you want to save the outputs of the workflow. A description of the output - including help to perform quality control of your data and interpretation of your results - are given in the following application-specific chapters.
