Create a custom Trim adapter list (optional)
All workflows included in the plugin start by removing all remaining adapters present on the reads using the "Remove read-through adapters" from the Trim Reads tool. For workflows for DNA and RNAscan applications, this means that the reads are annotated with the UMI information before the trimming removes them, as shown in figure 5.15.
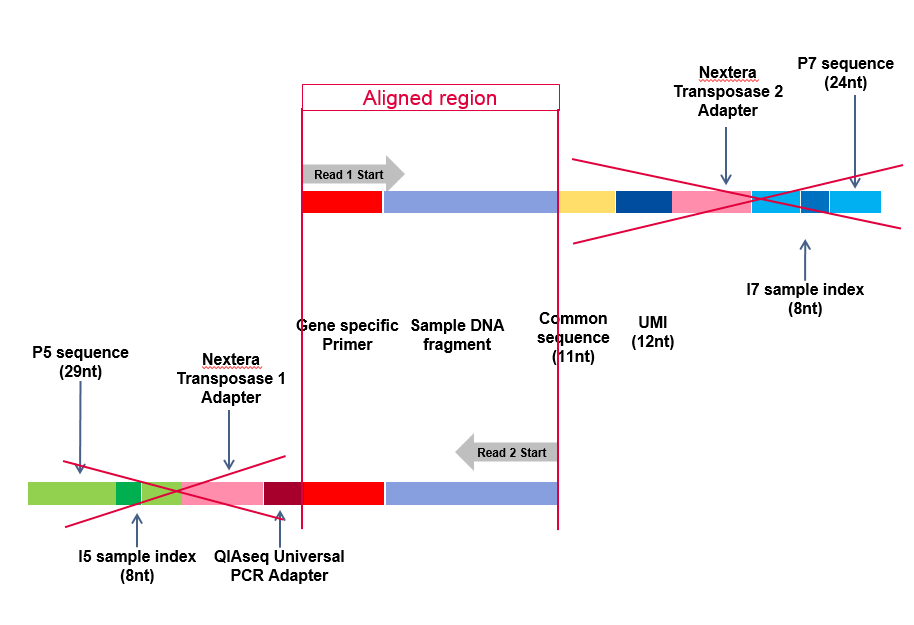
Figure 5.15: Adapters removed automatically from the reads by the Trim Sequences tool included in the workflow. This example shows reads sequenced with the UMI technology and an Illumina sequencer.
Despite the automatic trimming, remnants of various adapters and polyA and polyG sequences would hinder the mapping of the reads to a reference sequence if left on the reads. These pre-defined sequences are trimmed using a trim adapter list included in the reference data associated with each workflow application. Figure 5.16 shows all of the sequences potentially included in the different application-specific trim adapter list available in the CLC_References folder in the Navigation Area once the Reference Data Sets have been downloaded.
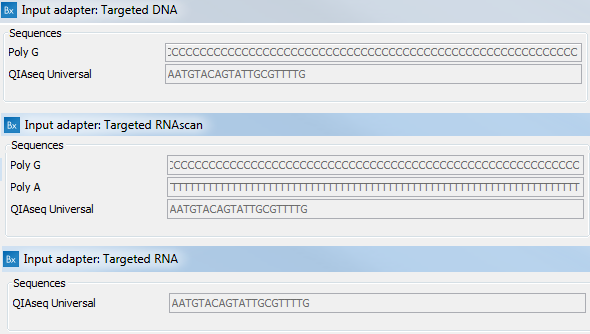
Figure 5.16: Application-specific templates for trim adapter lists.
In some particular cases, such as working with data of low quality, asymmetric read lengths, mate-paired reads and single reads, as well as when working with very large inserts length, it is recommended that you specify a trim adapter list that is more detailed than the one included in the reference data used by the analysis workflow. To find out if you need to run a custom workflow with a detailed trim adapter list, run the workflow once and check the report generated by the Trim Reads tool, and in particular the last section named "5 Automatic adapter read-through trimming" (figure 5.17). If that section shows no detected adapter, you should run the workflow again with a new Trim adapter list that contains the specific adapters of the sequencing technology used to generate the reads.
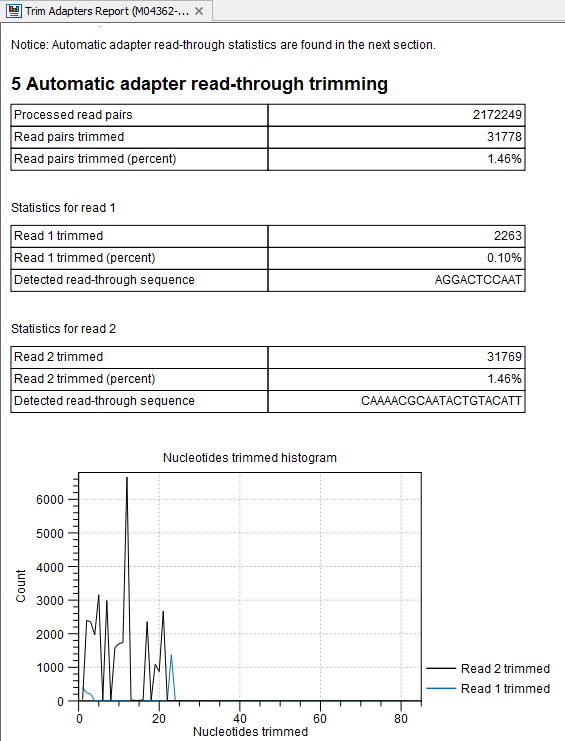
Figure 5.17: The Trim Adapters report indicates detected read-through sequences. If this section is empty, we recommend using a custom Trim Adapter list.
- Learn how to generate a Trim adapter list: http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Creating_new_Trim_adapter_list.html.
- Remember to add the adapters that were initially in the application specific template:
- for Targeted DNA applications: a poly G sequence and the QIAseq Universal adapter
- for Targeted RNAscan applications: a poly A sequence, a poly G sequence and the QIAseq Universal adapter
- for Targeted RNA applications: the QIAseq Universal adapter
- Learn how to create custom reference sets: http://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Custom_Sets.html.
