Filter Immune Repertoire
The Filter Immune Repertoire tool can be used to restrict clonotypes to only a specific subset, for example, only productive clonotypes, or clonotypes with a specific chain.
To run Filter Immune Repertoire go to the Toolbox and select:
Tools | QIAseq Panel Expert Tools (![]() ) | Immune Repertoire Expert Tools (
) | Immune Repertoire Expert Tools (![]() ) | Filter Immune Repertoire (
) | Filter Immune Repertoire (![]() )
)
This opens a dialog where a TCR clonotypes element (![]() ) or BCR clonotypes element (
) or BCR clonotypes element (![]() ) can be selected. Click Next to navigate through the different dialogs and configure the execution (see figures 8.11, 8.12, and 8.13).
) can be selected. Click Next to navigate through the different dialogs and configure the execution (see figures 8.11, 8.12, and 8.13).
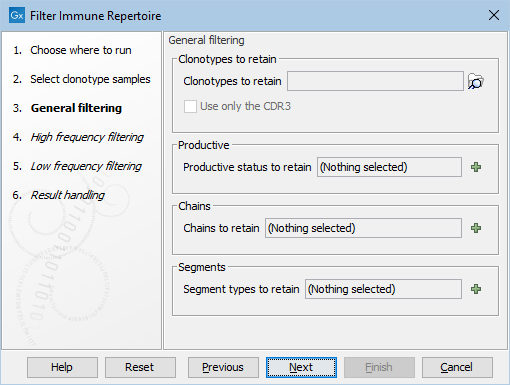
Figure 8.11: The default settings in the General filtering dialog.
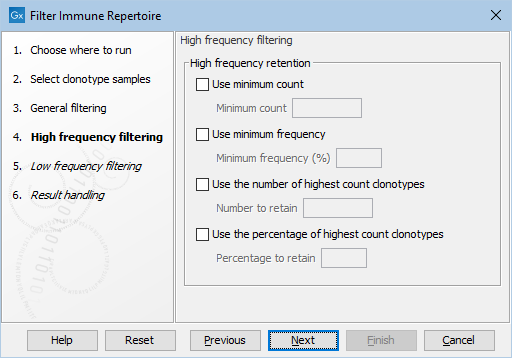
Figure 8.12: The default settings in the High frequency filtering dialog.
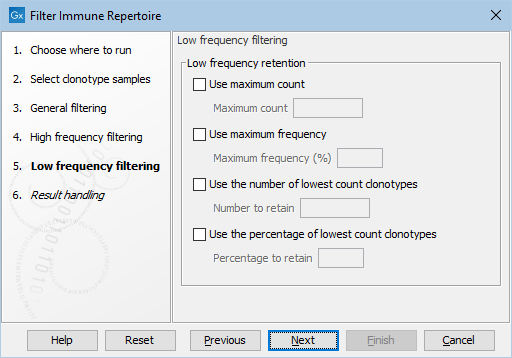
Figure 8.13: The default settings in the Low frequency filtering dialog.
Multiple filter options can be selected to obtain the desired output. Note that the filters are applied independently.
- Clonotypes to retain. Retain clonotypes that are found in all provided TCR (
 ) or BCR (
) or BCR ( ) elements. If left empty, no filter is applied.
) elements. If left empty, no filter is applied.
- Use only the CDR3. When comparing the clonotypes in the input with those in the elements from Clonotypes to retain, only the CDR3 is used if this is ticked. Otherwise, the V and J segments together with the CDR3 are used to determine if two clonotypes are the same.
- Productive status to retain. A combination of 'Productive', 'Out of frame' and 'Premature stop codon' can be chosen and only the clonotypes with the respective productive status will be retained. If left empty, no filter is applied.
- Chains to retain. A combination of 'TRA', 'TRB', 'TRG' and 'TRD' for TCR data, or 'IGH', 'IGK' and 'IGL' for BCR data, can be chosen and only the clonotypes with the respective chains will be retained. If left empty, no filter is applied.
- Segment types to retain. A combination of 'V', 'D', 'J' and 'C' can be chosen and only the clonotypes that have identified segments for all respective segment types will be retained. This means that, for example, if 'D' is chosen, only chains for which the D segment is used will be retained, and for those chains, only the clonotypes for which the identification of the D segment was successful will be retained. If left empty, no filter is applied.
- High frequency retention. The following filters for removing clonotyoes with low frequencies can be enabled:
- Use minimum count. Retain clonotypes with a count greater than or equal to Minimum count.
- Use minimum frequency. Retain clonotypes with a frequency greater than or equal to Minimum frequency (%).
- Use the number of highest count clonotypes. Retain Number to retain clonotypes that have highest frequency.
- Use the percentage of highest count clonotypes. Retain Percentage to retain percentage of clonotypes that have highest frequency.
- Retain clonotypes with low frequency. These filters are used to remove clonotypes that have high frequencies. Clonotypes can be filtered in multiple ways:
- Use maximum count. Retain clonotypes with a count less than or equal to Minimum count.
- Use maximum frequency. Retain clonotypes with a frequency less than or equal to Minimum frequency (%).
- Use the number of lowest count clonotypes. Retain Number to retain clonotypes that have lowest frequency.
- Use the percentage of lowest count clonotypes. Retain Percentage to retain percentage of clonotypes that have lowest frequency.
- Recalculate frequencies. If ticked, frequencies in the output clonotypes are recalculated such that they add up to 100%. Otherwise, the original frequencies found in the input are used.
It can be useful to recalculate frequencies when removing noise (for example, removing clonotypes with a count of 1), but if a subset of clonotpes is created for the purpose of comparing clonotypes between samples, it might be more relevant to preserve the original frequencies.
