Identify Rare Disease Causing Mutations in Family of Four (WES)
This workflow has been deprecated and will be retired in a future version of the software. It has been moved to the Legacy Workflows (![]() ) folder of the Toolbox, and its name has "(legacy)" appended to it. If you have concerns about the future retirement of this workflow, please contact QIAGEN Bioinformatics Support team at ts-bioinformatics@qiagen.com.
) folder of the Toolbox, and its name has "(legacy)" appended to it. If you have concerns about the future retirement of this workflow, please contact QIAGEN Bioinformatics Support team at ts-bioinformatics@qiagen.com.
You can use the Identify Rare Disease Causing Mutations in a Family of Four (WES) (legacy) template workflow to identify de novo and compound heterozygous variants from an extended family of four, where the fourth individual is not affected.
To run the Identify Rare Disease Causing Mutations in a Family of Four (WES) (legacy) workflow, go to:
Toolbox | Template Workflows | Legacy Workflows (![]() ) | Identify Rare Disease Causing Mutations in a Family of Four (WES) (legacy) (
) | Identify Rare Disease Causing Mutations in a Family of Four (WES) (legacy) (![]() )
)
- Double-click on the Identify Rare Disease Causing Mutations in a Family of Four (WES) (legacy) workflow to start the analysis. If you are connected to a server, you will first be asked where you would like to run the analysis.
- The sequencing reads from the different family members are specified one at a time in successive dialogs (figure 27.45).
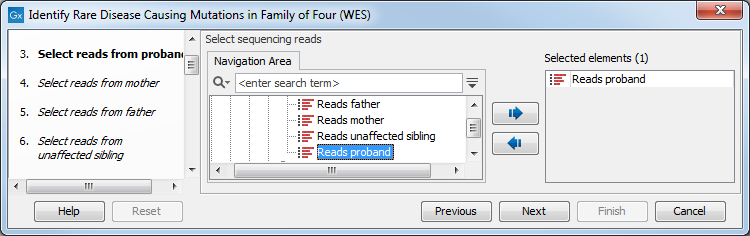
Figure 27.31: Specify the sequencing reads for the appropriate family member. - Select the targeted region file (figure 27.46).
The targeted region file is a file that specifies which regions have been sequenced, when working with whole exome sequencing or targeted amplicon sequencing data. This file is something that you must provide yourself, as this file depends on the technology used for sequencing. You can obtain the targeted regions file from the vendor of your targeted sequencing reagents.
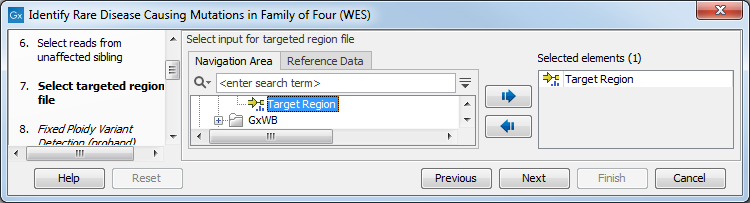
Figure 27.32: Select the targeted region file you used for sequencing. - Select which reference data set should be used to identify causal inherited variants (figure 27.47).
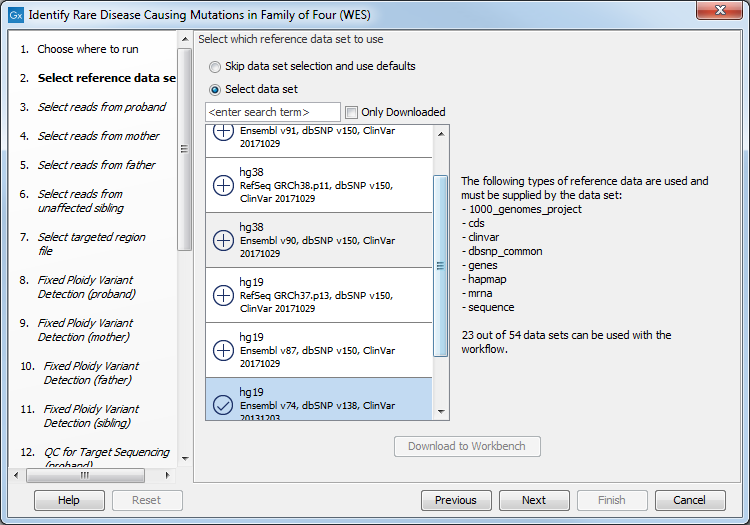
Figure 27.33: Choose the relevant reference Data Set to identify causal inherited variants. - Specify the Hapmap populations that should be used for filtering out variants found in Hapmap for each family member successively, with de novo and recessive being the proband variants (figure 27.48).
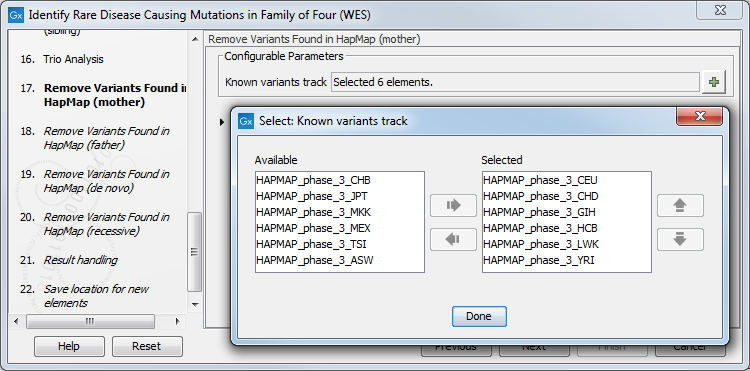
Figure 27.34: Select the relevant Hapmap population(s). - Specify the parameters for the Fixed Ploidy Variant Detection tool for each family member successively (figure 27.49).
The parameters used by the Fixed Ploidy Variant Detection tool can be adjusted. We have optimized the parameters to the individual analyses, but you may want to tweak some of the parameters to fit your particular sequencing data. A good starting point could be to run an analysis with the default settings.
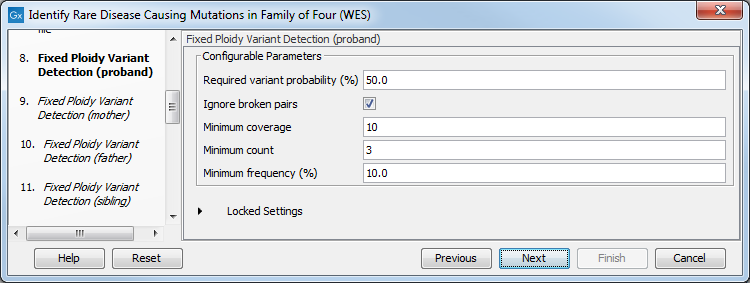
Figure 27.35: Specify the parameters for the Fixed Ploidy Variant Detection tool.The parameters that can be set are:
- Required variant probability is the minimum probability value of the 'variant site' required for the variant to be called. Note that it is not the minimum value of the probability of the individual variant. For the Fixed Ploidy Variant detector, if a variant site - and not the variant itself - passes the variant probability threshold, then the variant with the highest probability at that site will be reported even if the probability of that particular variant might be less than the threshold. For example if the required variant probability is set to 0.9 then the individual probability of the variant called might be less than 0.9 as long as the probability of the entire variant site is greater than 0.9.
- Ignore broken pairs: When ticked, reads from broken pairs are ignored. Broken pairs may arise for a number of reasons, one being erroneous mapping of the reads. In general, variants based on broken pair reads are likely to be less reliable, so ignoring them may reduce the number of spurious variants called. However, broken pairs may also arise for biological reasons (e.g. due to structural variants) and if they are ignored some true variants may go undetected. Please note that ignored broken pair reads will not be considered for any non-specific match filters.
- Minimum coverage: Only variants in regions covered by at least this many reads are called.
- Minimum count: Only variants that are present in at least this many reads are called.
- Minimum frequency: Only variants that are present at least at the specified frequency (calculated as 'count'/'coverage') are called.
- Specify the parameters for the QC for Targeted Sequencing tool for each family member successively (figure 27.50).
When working with targeted data (WES or TAS data), quality checks for the targeted sequencing is included in the workflows. Again, you can choose to use the default settings, or you can choose to adjust the parameters.
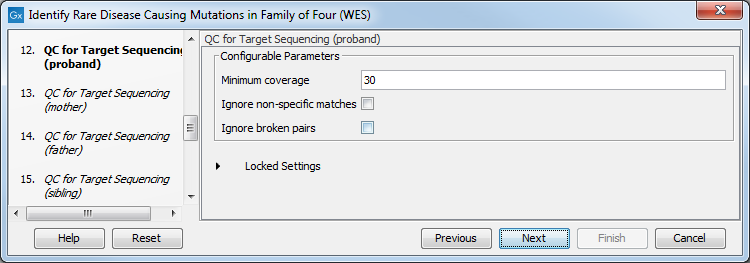
Figure 27.36: Specify the parameters for the QC for Targeted Sequencing tool.The parameters that can be set are:
- Minimum coverage provides the length of each target region that has at least this coverage.
- Ignore non-specific matches: reads that are non-specifically mapped will be ignored.
- Ignore broken pairs: reads that belong to broken pairs will be ignored.
- Specify the affected sibling's gender (figure 27.51).
Figure 27.37: Specify the proband's gender. - In the last wizard step you can check the selected settings by clicking on the button labeled Preview All Parameters.
In the Preview All Parameters wizard you can only check the settings, and if you wish to make changes you have to use the Previous button from the wizard to edit parameters in the relevant windows.
- Choose to Save your results and click on the button labeled Finish.
Output from the Identify Rare Disease Causing Mutations in a Family of Four (WES) (legacy) workflow
The following outputs are generated:
- Read Mapping One for each family member. The reads mapped to the reference sequence.
- Variant Tracks One for each family member. The variants identified in each of the family members. The variant track can be opened in table view to see all information about the variants.
- Target Region Coverage One track for each individual. When opened in table format, it is possible to see a range of different information about the targeted regions, such as target region length, read count, and base count.
- Target Region Coverage Report One for each family member. The report consists of a number of tables and graphs that in different ways provide information about the mapped reads from each sample.
- Identified Compound Heterozygous Genes Proband Gene track with the identified putative compound heterozygous Variants in the proband. The gene track can be opened in table view to see the gene names.
- Gene List with de novo Variants Gene track with the identified putative compound heterozygous Variants in the proband. The gene track can be opened in table view to see the gene names.
- Gene List with recessive Variants Gene track with the identified recessive variants in the proband. The gene track can be opened in table view to see the gene names.
- De novo variants Variant track showing de novo variants in the proband. The variant track can be opened in table view to see all information about the variants.
- Recessive variants Variant track showing recessive variants in the proband. The variant track can be opened in table view to see all information about the variants.
- De novo Mutations Amino Acid Track
- Recessive Variants Amino Acid Track
- Track List This is a collection of tracks shown together in a view that makes it easy to compare information from the individual tracks, such as compare the identified variants with the read mappings and information from databases.
