Reference data for analyses on the cloud
When running workflows configured to use QIAGEN reference data, no transfer of that data will take place, and there is no need to explicitly copy it to your own AWS S3 account. Under certain conditions, no local copy of this reference data is needed either. This is described in more detail later in this section.
In cases where other reference data will be used, the following steps are recommended:
- Connect Input elements to all input channels that require reference data (figure 6.1).
When a workflow Input element is connected, you are presented with the option of using on-the-fly import. This allows you to select data in an AWS S3 bucket (figure 6.2).
- Upload reference data to AWS S3 before launching analyses.
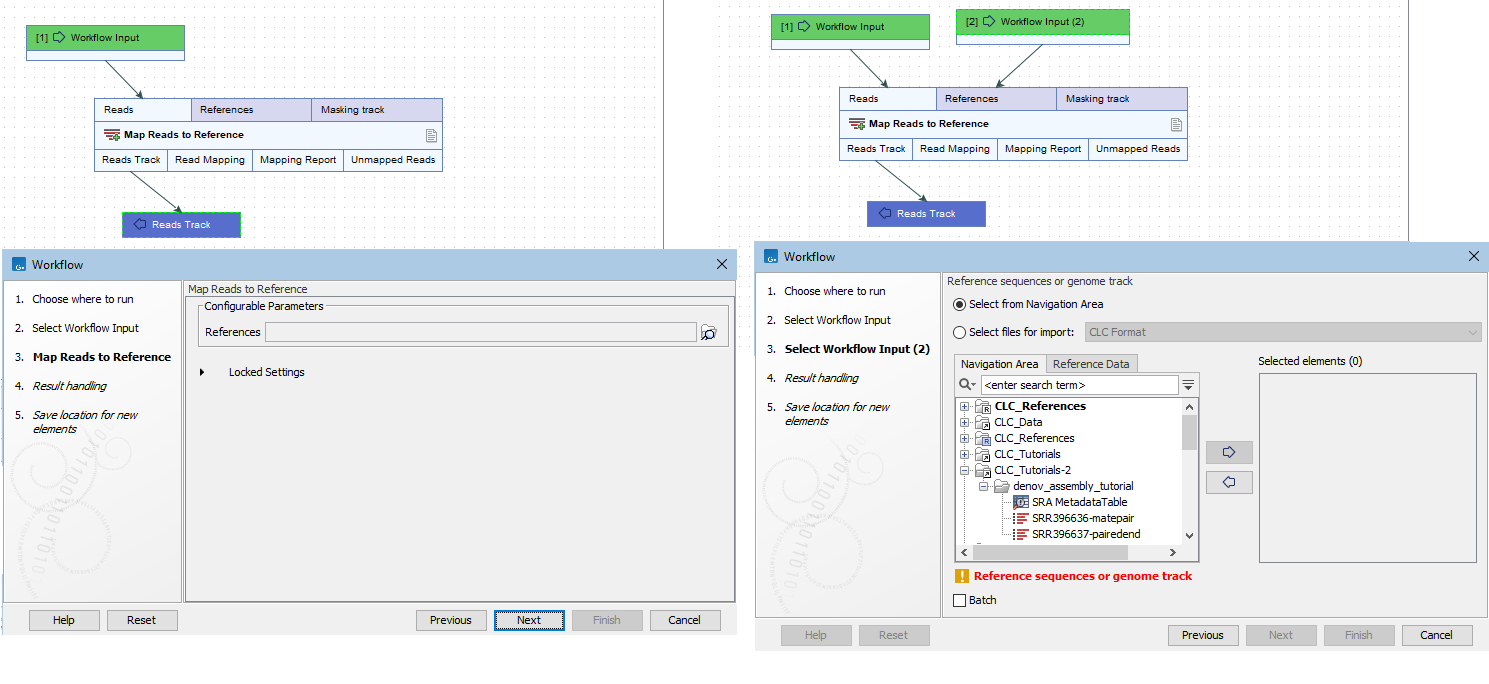
Figure 6.1: Using the workflow on the left, data for the References field can only be selected from a CLC Location. The workflow on the right has an Input element connected to the References input channel. Using that workflow, files can be selected from an AWS S3 bucket, or from other accessible places, including CLC Locations.
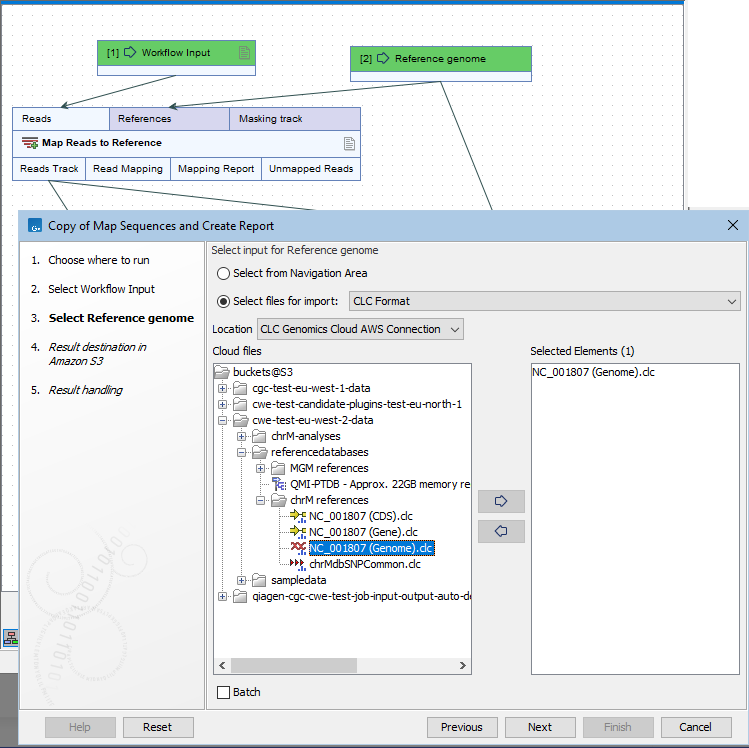
Figure 6.2: An Input element is connected to the References input channel. On-the-fly import of a CLC format file has been specified by selected the "Select files for on-the-fly import" option and "CLC Format" from the drop-down list of formats. The relevant AWS Connection has been selected from the drop-down list of locations. A CLC file was then selected for use as the reference genome.
QIAGEN reference data in workflows
QIAGEN reference data elements6.1 are already present in AWS S3 (figure 6.3) and thus do not need to be uploaded to your own S3 bucket when running workflows configured to refer to them. This includes many of the template workflows delivered with the software, and thus also workflows derived from those.
When the conditions listed below are met, there is also no need for a local copy of QIAGEN reference data when launching workflows to run on the cloud.
- All reference data parameters must be configured with a single QIAGEN reference data element and/or configured with a workflow role that is specified in one or more QIAGEN reference sets.
Configuring workflows with roles for reference data is described at https://resources.qiagenbioinformatics.com/manuals/clcgenomicsworkbench/current/index.php?manual=Configuring_Workflow_Input_elements.html.
- All workflow parameters referring to QIAGEN reference data are locked.
Where reference data elements need to be specified when launching, those elements, by definition, must be unlocked, and so a locally accessible copy of the relevant reference data will be needed. This is the case, for example, for many QIAseq analysis workflows provided by the Biomedical Genomics Analysis plugin, where there are drop-down menus for selecting target region and target primer sets. If QIAGEN reference elements are selected, they are not copied to S3 from your system. Instead, the copy provided by QIAGEN in AWS S3 will be used for analyses run on the cloud.
- Workflows are being submitted via a CLC Workbench.
When submitting workflows that make use of QIAGEN reference data via a CLC Server, the reference data elements must be present in the CLC Server CLC_References location. Any QIAGEN reference elements selected will, however, not be copied to S3 from your system. The copy provided in AWS S3 by QIAGEN will be used for analyses run on the cloud.
Note: To view track lists that refer to reference data elements, those elements must be available locally.
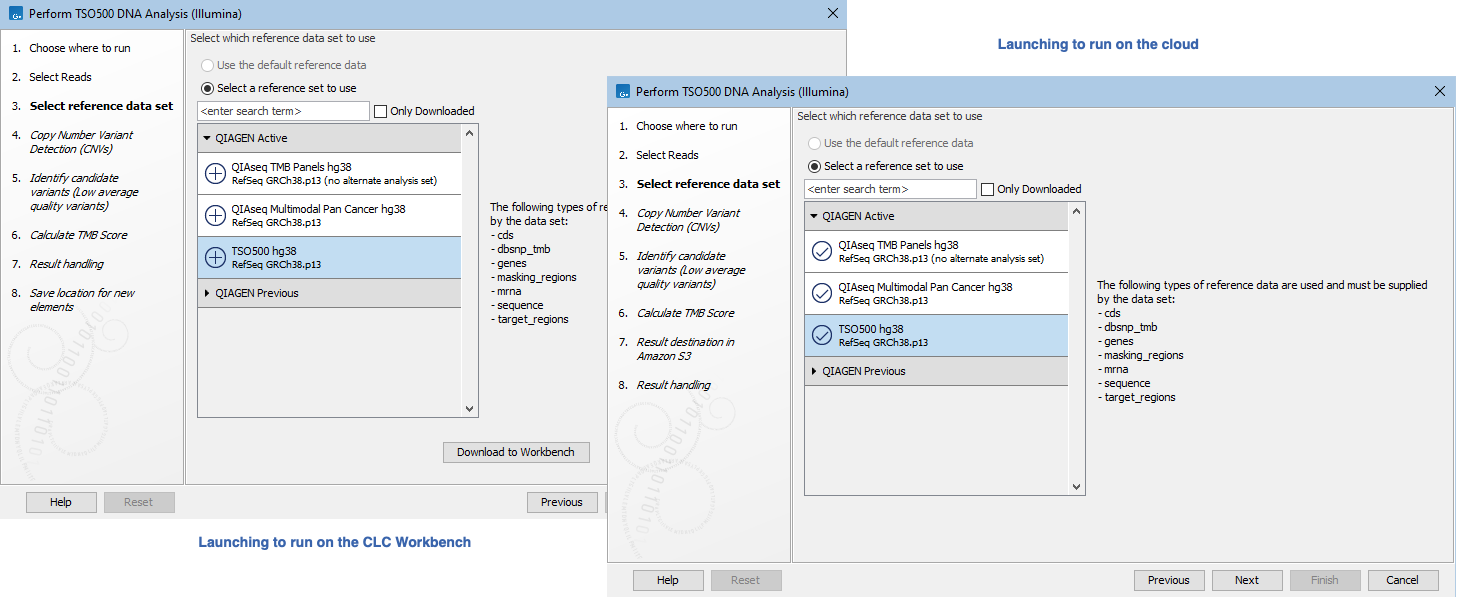
Figure 6.3: QIAGEN Reference Sets do not need to be available locally (left hand image) for them to be available when launching a workflow to run on the cloud (right hand image).
Footnotes
- ... elements6.1
- "QIAGEN reference data" refers to data sets or data elements provided by QIAGEN, available from under the QIAGEN references tab of the Reference Data Manager. Template workflows, delivered with the software, are commonly configured to use QIAGEN reference data.
