Setting up AWS resources
We recommend using the cgc-standard.json CloudFormation template, described below, to set up the standard resources needed for a CLC Genomics Cloud setup. Some configuration can be done when using this template. Additional AWS Batch queues can be added using another CloudFormation template afterwards. That template provides more configuration options (see Adding more AWS Batch queues for CLC jobs).
Below the description of the resources established using the cgc-standard.json CloudFormation template are detailed instructions of how to use the template, as well as information about the AWS IAM users created using the template.
Creating stacks using the AWS CloudFormation console is described in the AWS documentation at https://docs.aws.amazon.com/AWSCloudFormation/latest/UserGuide/cfn-console-create-stack.html.
Overview of standard CLC Genomics Cloud infrastructure on AWS
The cgc-standard.json CloudFormation template defines the resources needed for a CLC Genomics Cloud, which include:
- Three AWS Batch queues The queues are named
cgc-small,cgc-medium, andcgc-large. When a user launches a job to run on the cloud from their CLC Workbench, they select a queue to send the job to from a drop down list of the available queues. Details of each of these queues are provided in Standard AWS Batch queues for CLC Genomics Cloud. - An S3 bucket for system files The name of this bucket begins with "cgc-system-".
This bucket is used by the CLC software for system files, including read mapper indexes. It is not intended for storing sample data or results. Details about the retention policies on this bucket are provided in System file retention policies.
The system bucket will not be accessible for browsing from a CLC Workbench and cannot be browsed by non-admin users logged into the CLC Server web client. It can be browsed by CLC Server admin users logged into the web client.
Note: S3 buckets for holding input data and results need to be created directly in AWS. They are not created by CloudFormation templates provided by QIAGEN.
- Two AWS IAM users One with properties supporting submission of analyses, and the other allowing only access to AWS S3 buckets. This is described in more detail below.
When working with a CLC Workbench, access to the AWS Batch queues is determined by the access rights of the AWS IAM user configured in the AWS connection. With a CLC Server, access to AWS Batch queues can be fine tuned by setting group permissions on cloud presets, using the web administrative interface of the CLC Server, as described in Configuring cloud presets.
Creating CLC Genomics Cloud infrastructure on AWS
To set up the standard infrastructure on AWS for handling CLC jobs:
- Log into the AWS console as a user with privileges that allow the infrastructure described above to be created.
- Set the region to the one the AWS resources should be established in.
- Copy the URL below:
https://qiagen-clc-genomics-cloud-formation.s3.eu-central-1.amazonaws.com/cgc-standard.json
- Go to the CloudFormation service and click on Create stack.
- In the Create stack step, keep "Choose an existing template" selected. In the "Specify template" step, keep the "Amazon S3 URL" option selected and paste the CloudFormation template URL you just copied into the "Amazon S3 URL" field.
- In the next step, specify a stack name and add a unique ID in the Parameters section.
- Review the max vCPUs setting for each queue.
By default, the values allow for up to 10 EC2 instances to be launched. I.e. up to 10 jobs can be run in parallel. This value can be increased or decreased. We recommend it is not decreased below the number of cores designated for each job. These values can be found in the setting details provided in Standard AWS Batch queue settings.
- Review the Disk size setting for each queue.
This value refers to the space available for each job when it is running.
- Specify the URL for custom plugin repository if you have custom CLC plugins providing tools that should run on the cloud.
See Making custom plugins available for cloud analyses for the requirements for a custom plugin location.
This field should be left blank if you require access only to plugins and modules distributed by QIAGEN. These are the plugins and modules listed in CLC Workbench and CLC Server Plugin Manager, or on the QIAGEN website:
https://digitalinsights.qiagen.com/products-overview/plugins/ - Step through the rest of the stack creation. No other settings require configuration.
- When prompted, agree to the AWS conditions and click on the Submit button.
AWS IAM users
When stack creation is complete, go to the Outputs tab of the main stack to find the credentials for the AWS IAM users created (figure 2.1).
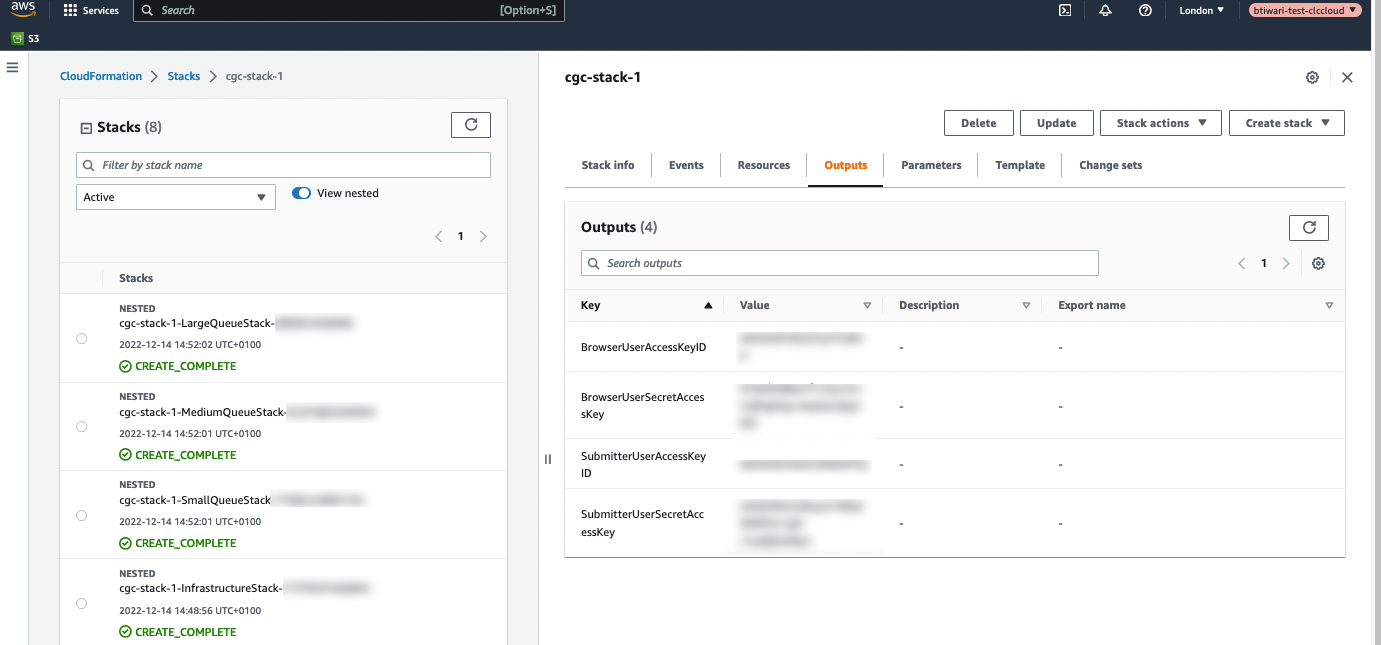
Figure 2.1: The credentials for the AWS IAM users created using the CloudFormation template are listed under the Outputs tab for the stack.
AWS Connections using the "SubmitterUser" (CgcSubmitterUser-<EnvironmentId>) credentials allow CLC analyses to be submitted an AWS Batch queue for analysis. This user also has full access to AWS S3, read access to CloudWatch logs, and can list CloudFormation resources.
AWS Connections using the "BrowserUser" (CgcBrowserUser-<EnvironmentId>) credentials support listing S3 buckets and accessing bucket contents. Jobs cannot be submitted to run on AWS using these credentials.
AWS IAM user credentials are entered in AWS Connections in CLC Workbenches or the CLC Server, described in Configuring the AWS connection in the Workbench and Configuring the AWS Connection in the CLC Server, respectively.
The full policy for each user can be viewed in the Identity and Access Management (IAM) area of the AWS Console.
AWS S3 buckets for storing input data and results
One or more AWS S3 buckets must be created for holding input data and results. These buckets must be created in the same AWS account and region that the AWS Batch queues were established in. Please refer to AWS documentation for details:
https://docs.aws.amazon.com/AmazonS3/latest/userguide/create-bucket-overview.html.
Note: The prefix cgc-system- should be considered reserved. Buckets given names starting with this term will not be visible in CLC Workbenches.
