Metadata
Data elements may be associated with metadata such as information about samples, patients, families, cohortes etc. Metadata is used in a variety of ways in the Workbench, including searching and filtering data elements and annotating phylogenetic trees. The metadata system is intended to help you keep track of your data elements, and how they relate to your samples, without having to resort to elaborate and hard-to-maintain naming conventions or folder structures. For instance, metadata is automatically transferred from input to output elements when an analysis tool is executed.
The Workbench handles metadata in tabular form. A Metadata Table represents a homogeneous collection of external entities, typically samples. Within such a Metadata Table of samples, each row represents a particular sample, and each column represents a property of the samples in the collection. The Workbench does not force any particular interpretation of metadata properties, so you are free to use them to suit your purpose. In the example Metadata Table shown in figure 3.8, the cell containing the value 4 means that the sample with ID ETC-006 has the value 4 for its 'Batch' property. To you, that could e.g. mean that the sample was analyzed with batch 4 chemistry.
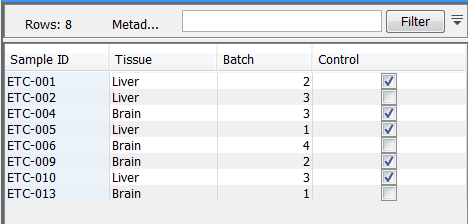
Figure 3.8: A simple Metadata Table.
Any number of Metadata Tables may be created in the Workbench, each with its own set of columns. A data element may be associated with at most one row in each Metadata Table (but you can have references in more metadata tables). Many data elements contain information about a particular sample, and so they may be associated with the same row in a Metadata Table representing samples. For cross-sample analysis results, the association to a sample row would not be unique; such results may instead be associated with a single row in a Metadata Table representing suitable multi-sample entities such as families or cohortes.
Subsections
- Setting up Metadata Tables
- Editing the metadata structure
- Importing metadata columns
- Editing metadata rows
- Importing metadata rows
- Associating data elements with metadata
- Finding data elements based on metadata
- Viewing metadata associations
- Exporting metadata
