Over-representation analyses
- Enriched 5-mer distribution
- The 5-mer analysis examines the enrichment of penta-nucleotides. The enrichment of 5-mers is calculated as the ratio of observed and expected 5-mer frequencies. The expected frequency is calculated as product of the empirical nucleotide probabilities that make up the 5-mer. (Example: given the 5-mer = CCCCC and cytosines have been observed to 20% in the examined sequences, the 5-mer expectation is
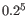 ). Note that 5-mers that contain ambiguous bases (anything different from A/T/C/G) are ignored. This analysis calculates the absolute coverage and enrichment for each 5-mer (observed/expected based on background distribution of nucleotides) for each base position, and plots position vs enrichment data for the top five enriched 5-mers (or fewer if less than five enriched 5-mers are present). It will reveal if there is a bias at certain positions along the read length. This may originate from non-trimmed adapter sequences, poly A tails and more.
). Note that 5-mers that contain ambiguous bases (anything different from A/T/C/G) are ignored. This analysis calculates the absolute coverage and enrichment for each 5-mer (observed/expected based on background distribution of nucleotides) for each base position, and plots position vs enrichment data for the top five enriched 5-mers (or fewer if less than five enriched 5-mers are present). It will reveal if there is a bias at certain positions along the read length. This may originate from non-trimmed adapter sequences, poly A tails and more.
- Sequence duplication levels
- The duplicated sequences analysis identifies sequences that have been sequenced multiple times. In order to achieve reasonable performance, not all input sequences are analyzed. Instead a sequence-dictionary of 250 000 sequences is used, whose entries are sampled evenly from the input dataset. Please note that if you select multiple sequence lists as an input, they will all be considered one data set for this analysis. You can use the batch mode to generate separate reports for individual sequence lists. As soon as a sequence makes it into the dictionary (which is a random process), it is tracked for duplicates until all sequences have been examined. Because all current sequencing techniques tend to report decreasing quality scores for the 3' ends of sequences, there is a risk that duplicates are NOT detected, just because of sequencing errors near their 3' ends. Therefore, the identity of two sequences is calculated using only the first 50nt from the 5' end. The results of this analysis is a plot correlating duplication counts with the number of sequences that featured that duplicate-count. For example, if the dictionary contains 10 sequences and each sequence was seen exactly once, then the table will contain only one row displaying: duplication-count=1 and sequence-count=10. A high level of duplication may indicate enrichment bias (e.g. introduced by PCR amplification). Note: due to space restrictions the corresponding bar-plot shows only bars for duplication-counts of x=[0-100]. Bar-heights of duplication-counts >100 are accumulated at x=100. Please refer to the table-report for a full list of individual duplication-counts.
- Duplicated sequences
- This results in a list of actual sequences most prevalently observed. The list contains a maximum of 25 (most frequently observed) sequences and is only present in the supplementary report.
