Standard import
Biomedical Genomics Workbench has support for a wide range of bioinformatic data such as molecules, sequences, alignments etc. See a full list of the data formats in List of data formats.
These data can be imported through the Import dialog, using drag/drop or copy/paste as explained below.
For import of NGS data, please see Import high-throughput sequencing data For import of tracks, please see Import tracks.
Import using the import dialog
To start the import using the import dialog:
click Import (![]() ) in the Toolbar | Standard Import
) in the Toolbar | Standard Import
This will show a dialog similar to figure 6.1. You can change which kind of file types that should be shown by selecting a file format in the Files of type box.
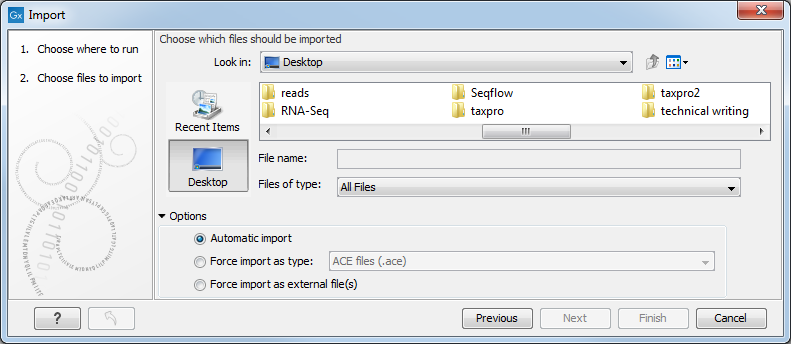
Figure 6.1: The import dialog.
Next, select one or more files or folders to import and click Next to select a place for saving the result files. If you import one or more folders, the contents of the folder is automatically imported and placed in that folder in the Navigation Area. If the folder contains subfolders, the whole folder structure is imported.
In the import dialog (figure 6.1), there are three import options:
- Automatic import
- This will import the file and Biomedical Genomics Workbench will try to determine the format of the file. The format is determined based on the file extension (e.g. SwissProt files have .swp at the end of the file name) in combination with a detection of elements in the file that are specific to the individual file formats. If the file type is not recognized, it will be imported as an external file. In most cases, automatic import will yield a successful result, but if the import goes wrong, the next option can be helpful:
- Force import as type
- This option should be used if Biomedical Genomics Workbench cannot successfully determine the file format. By forcing the import as a specific type, the automatic determination of the file format is bypassed, and the file is imported as the type specified.
- Force import as external file
- This option should be used if a file is imported as a bioinformatics file when it should just have been external file. It could be an ordinary text file which is imported as a sequence.
Import using drag and drop
It is also possible to drag a file
from e.g. the desktop into the Navigation Area of Biomedical Genomics Workbench. This
is equivalent to importing the file using the Automatic import
option described above. If the file type is not recognized, it will
be imported as an external file.
Import using copy/paste of text
If you have e.g.
a text file or a browser displaying a sequence in one of the formats
that can be imported by Biomedical Genomics Workbench, there is a very easy way to
get this sequence into the Navigation Area:
Copy the text from the text file or browser |
Select a folder in the Navigation Area |
Paste (![]() )
)
This will create a new sequence based on the text copied. This operation is equivalent to saving the text in a text file and importing it into the Biomedical Genomics Workbench.
If the sequence is not formatted, i.e. if you just have a text like this: "ATGACGAATAGGAGTTCTAGCTA" you can also paste this into the Navigation Area.
Note! Make sure you copy all the relevant text - otherwise Biomedical Genomics Workbench might not be able to interpret the text.
Subsections
